Integrative Genomic, Transcriptomic and Epigenomic Analysis Reveals cis-regulatory Contributions to High-altitude Adaptation in Tibetan Pigs
- PMID: 40644385
- PMCID: PMC12290512
- DOI: 10.1093/molbev/msaf169
Integrative Genomic, Transcriptomic and Epigenomic Analysis Reveals cis-regulatory Contributions to High-altitude Adaptation in Tibetan Pigs
Abstract
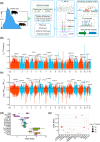
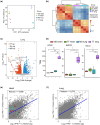
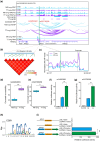
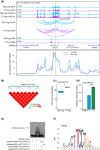
The Qinghai-Tibet Plateau, characterized by its extreme environmental conditions, presents significant challenges to life, making it an ideal region for studying adaptation and evolution. Tibetan pigs, known for their high genetic diversity and exceptional adaptability to high altitudes, serve as excellent models for investigating high-altitude adaptation. While previous studies have extensively identified genetic determinants associated with high-altitude adaptation, the molecular mechanisms, particularly cis-regulatory patterns, remain poorly understood. Here, we conducted a selective sweep analysis using 484 genomes from Chinese and Western pig breeds across various altitudes, revealing 38.56 Mb of genomic regions under selection in Tibetan pigs. Enrichment analysis identified the lung as the primary functional tissue involved in high-altitude adaptation, supported by tissue-specific transcriptional and regulatory patterns observed between Tibetan and Meishan pigs (low altitude). By integrating genomic, RNA-seq, ATAC-seq, and H3K27ac HiChIP data, we constructed comprehensive enhancer-promoter regulatory maps of candidate genes and pinpointed promising genetic determinants associated with high-altitude adaptation, including SNPs in EPAS1, KLF13, SPRED1, and CFD. These loci were predicted to influence chromatin accessibility and the interactions of regulatory elements, with altered binding strength of relevant transcription factors. Further in vitro experiments confirmed that these loci function as allele-specific enhancers, modulating the expression of target genes. Our findings elucidate the regulatory basis of high-altitude adaptation in Tibetan pigs and provide valuable insights for exploring hypoxia-related diseases in livestock and humans.
Keywords: H3K27ac HiChIP; Tibetan pigs; high-altitude adaptation; lung tissue; multi-omics integration.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest: The authors declare no conflict of interest.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

