Cypin regulates K63-linked polyubiquitination to shape synaptic content
- PMID: 40644549
- PMCID: PMC12248297
- DOI: 10.1126/sciadv.ads5467
Cypin regulates K63-linked polyubiquitination to shape synaptic content
Abstract
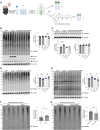
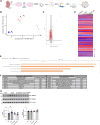
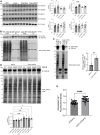
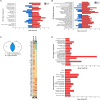
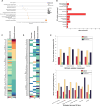
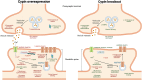
An open question in neuroscience is how protein posttranslational modifications regulate synaptic site targeting. Polyubiquitination plays a role in proteasome-mediated protein turnover; however, additional functions for specific types of polyubiquitin linkages have been identified. One type of polyubiquitination, K63-polyubiquitin (K63-polyUb), has been studied for its role in signal transduction within the context of cancer, but little has been done to uncover its role regarding neuronal and synaptic function. Here, we report an emerging function for the cytosolic PSD-95 interactor, cypin, in the regulation of synaptic content by K63-polyUb during neuronal development in vitro and in adult mice in vivo. We identify cypin-promoted K63-polyUb on postsynaptic proteins and also find an important role for cypin in presynaptic function. Our work demonstrates that cypin-promoted changes to K63-polyUb can regulate synaptic content and function on both sides of the synapse, adding important insight into basic mechanisms of neuronal signaling.
Figures


References
-
- Soykan T., Haucke V., Kuijpers M., Mechanism of synaptic protein turnover and its regulation by neuronal activity. Curr. Opin. Neurobiol. 69, 76–83 (2021). - PubMed
-
- Yi J. J., Ehlers M. D., Ubiquitin and protein turnover in synapse function. Neuron 47, 629–632 (2005). - PubMed
-
- Sheng M., Hoogenraad C. C., The postsynaptic architecture of excitatory synapses: A more quantitative view. Annu. Rev. Biochem. 76, 823–847 (2007). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

