UFMylation maintains tumor suppressor pVHL stability by activating the deubiquitinase BAP1
- PMID: 40644555
- PMCID: PMC12248380
- DOI: 10.1126/sciadv.adt8800
UFMylation maintains tumor suppressor pVHL stability by activating the deubiquitinase BAP1
Abstract
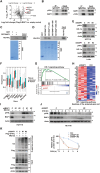
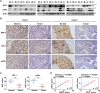
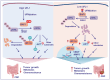
BRCA1-associated protein 1 (BAP1) can function as a tumor suppressor or oncogene depending on context, but its role in colorectal cancer (CRC) is not well understood. Here, we demonstrate that BAP1 suppresses CRC progression primarily by deubiquitinating and stabilizing von Hippel-Lindau tumor suppressor protein (pVHL). BAP1 undergoes covalent modification by ubiquitin-fold modifier 1 (UFM1) at Lys51, Lys61, Lys187, and Lys205, enhancing its interaction with pVHL and promoting pVHL stabilization. Loss of this modification through UFL1 depletion or reconstitution with a UFMylation-defective BAP1 mutant (4KR) impairs pVHL stabilization and promotes tumor progression in CRC cell line-based and patient-derived xenograft models. Clinically, down-regulation of UFL1 and BAP1 correlates with reduced pVHL level and poor prognosis in patients with CRC. These findings identify a previously unrecognized posttranslational mechanism regulating BAP1 activity and highlight UFMylation as essential for maintaining pVHL tumor-suppressive function. Targeting BAP1 UFMylation may represent a potential therapeutic strategy in CRC and other cancers with wild-type BAP1 and VHL.
Figures




References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). - PubMed
-
- Testa J. R., Cheung M., Pei J., Below J. E., Tan Y., Sementino E., Cox N. J., Dogan A. U., Pass H. I., Trusa S., Hesdorffer M., Nasu M., Powers A., Rivera Z., Comertpay S., Tanji M., Gaudino G., Yang H., Carbone M., Germline BAP1 mutations predispose to malignant mesothelioma. Nat. Genet. 43, 1022–1025 (2011). - PMC - PubMed
-
- Gerlinger M., Horswell S., Larkin J., Rowan A. J., Salm M. P., Varela I., Fisher R., McGranahan N., Matthews N., Santos C. R., Martinez P., Phillimore B., Begum S., Rabinowitz A., Spencer-Dene B., Gulati S., Bates P. A., Stamp G., Pickering L., Gore M., Nicol D. L., Hazell S., Futreal P. A., Stewart A., Swanton C., Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 46, 225–233 (2014). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

