Genotype and transcript processing of the tumour necrosis factor receptor TNFRSF1A in epithelial cells: implications for survival in cystic fibrosis
- PMID: 40645008
- PMCID: PMC12275981
- DOI: 10.1016/j.ebiom.2025.105848
Genotype and transcript processing of the tumour necrosis factor receptor TNFRSF1A in epithelial cells: implications for survival in cystic fibrosis
Abstract
Background: Cystic fibrosis is caused by mutations of the cystic fibrosis transmembrane conductance regulator, CFTR, an epithelial anion transport protein, responsible for, inter alia, sputum viscoelasticity in the lung. We previously identified the TNF receptor superfamily 1A TNFRSF1A (TNFR1) as a genetic modifier of CFTR function and disease severity in the CF twin and sibling study population. We aimed to replicate our findings in independent cohorts, assess the role of TNFR1 for patient survival and identify functional changes associated with TNFR1 polymorphisms.
Methods: We incorporated data from three independent long-term mono- and multicentric cohorts of people with cystic fibrosis (pwCF) to confirm the previously described association of TNFR1 with CFTR function and to extend our study to include survival data for our local cohort and a pan-European cohort of pwCF. We studied TNFR1 transcripts obtained from primary airway epithelia grown as air-liquid interface cultures to address possible mechanisms involved in up-stream and down-stream effects of TNFR1.
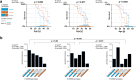
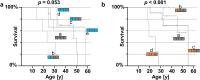
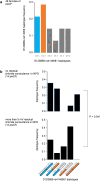
Findings: Survival differed by more than a decade when comparing carriers of contrasting TNFR1 genotypes among unrelated pwCF as well as among CF siblings pairs. The presence of the TNFR1 transcript variant TNFR1delEx2 in primary airway epithelia was associated with TNFR1 genotype.
Interpretation: The association of the TNFR1 transcript variant TNFR1delEx2 associates with the TNFR1 genotype, possibly mediating the genotype-survival association we found regarding TNFR1 genotype and patient survival in cystic fibrosis.
Funding: Supported by the German Ministry for Education and Research (BMBF) (82DZL009B1 to MAM and 82DZL002A1, to GH, BT, AMD, FS) and the Mukoviszidose Institut gGmbH (MI-2002, to LN, AMD, FS).
Keywords: Alternative transcript; Cystic fibrosis; Genetic association study; Patient survival; TNFRSF1A; Tumour necrosis factor receptor.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The following authors state no conflict of interest with respect to this manuscript’s content and in accordance with the ICMJE guidelines: Alexander Uden (AU); Inga Dunsche (ID); Longhua Feng (LF); Stephanie Tamm (ST); Silke Hedtfeld (SH); Gesa Stege (GS); Kirstin Jahn (KJ); Julia Kontsendorn (JK); Nadine Alfeis (NA); Ines Kühbandner (IK); Rebecca Minso (RM); Christian Dopfer (CD); Matthias Griese (MG); Gesine Hansen (GH); Frauke Stanke (FS). Sabina Janciauskiene (SJa); Simon Gräber (SG); Olaf Sommerburg (OS); Felix C. Ringshausen (FR); Lutz Nährlich (LN); Peter Braubach (PB); Marcus A Mall (MM); Burkhard Tümmler (BT) and Anna-Maria Dittrich (AMD) disclose institutional or personal grants, consulting fees or payment honoraria received within the past 36 months from: the European Union (FR); the German Centre for Lung Research-DZL (SJa); the Helmholtz centre for infection research (FR, BT); the German Innovation Fund (MM); the German CF foundation (SG, FR, LN); the Christiane-Herzog-Foundation (AMD); the Paul-Ehrlich-Institute (AMD); the German Kartagener Syndrome and Primary Ciliary Dyskinesia Patient Advocacy Group (FR); the Austrian Society for Rare Disease (AMD); the VW-Stiftung (BT); University Hospital Frankfurt (FR); University Hospital Hamburg (FR); University Dundee (FR); Social Court Cologne (FR); Vertex Pharmaceuticals Incorporated (SG, OS, FR, LN, MM, BT, AMD); I!DE Werbeagentur GmbH (FR); AstraZeneca (FR, PB); Arcturus (FR); Bayer (FR); Boehringer Ingelheim (FR, PB, MM); Chiesi GmbH (SG); Danone (OS), Enterprise Therapeutics (MM); Glaxo Smith-Kline (AMD); Grifols (FR, SJa); Hogrefe Publishing Company (AMD); InfectoPharm (FR); Insmed Germany (FR); Interkongress GmbH (FR); Kither Biotec (MM); Novartis (FR, AMD); Pari (OS, MM); Parion (FR); ReCode (FR); Sanofi (FR); Splisense (MM); Zambon (FR). The following authors participate on data safety monitoring or advisory boards of: Boehringer Ingelheim (FR, MM); Chiesi (SG, FR); Enterprise Therapeutics (MM); Grifols (FR); Insmed (FR); Kither Biotec (MM); Pari (MM); Trial Steering Committee for CF STORM (LN); Vertex Pharmaceutical Incorporated (SG, OS, BT). The following authors serve a leadership or fiduciary role in other board, society, committee or advocacy group: Coordinator of the ERN-LUNG Bronchiectasis Core Network (FR); Chair of the German Bronchiectasis Registry PROGNOSIS (FR); Member of the SteerCo of the European Bronchiectasis Registry EMBARC (FR); Member of the SteerCo of the European Nontuberculous Mycobacterial Pulmonary Disease Registry EMBARC-NTM (FR); Co-Speaker of the Medical Advisory Board of the German Kartagener Syndrome and PCD Patient Advocacy Group (FR); PI of the German Centre for Lung Research (FR); Member of the Protocol Review Committee of the PCD-CTN (FR); Medical lead of the German CF-registry (LN); Pharmacovigilance study manager of the European Cystic Fibrosis Society Patient Registry (LN); European Respiratory Society (MM); Fellow of the European Respiratory Society (MM); Christiane-Herzog-Foundation (BT); German CF patient advocacy board (AMD): German CF Clinical Trial Network ExecCommittee (AMD); European Cystic Fibrosis Society Clinical Trials network ExecCommittee (AMD). The consortium of CF teams was established through outreach efforts, during which caregivers of individuals with cystic fibrosis were contacted via telephone. Caregivers, physicians and other CF professionals who agreed to participate contributed survival data of participants of EUCFTSib and those contributors are listed here as CF teams from the European CF twin and sibling study.
Figures

References
-
- Chen G., Goeddel D.V. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. - PubMed
-
- Silke J. The regulation of TNF signalling: what a tangled web we weave. Curr Opin Immunol. 2011;23:620–626. - PubMed
-
- Gómez M.I., Lee A., Reddy B., et al. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat Med. 2004;10:842–848. - PubMed
-
- Gómez M.I., O’Seaghdha M., Magargee M., Foster T.J., Prince A.S. Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J Biol Chem. 2006;281:20190–20196. - PubMed
-
- Classen A., Kalali B.N., Schnopp C., et al. TNF receptor I on human keratinocytes is a binding partner for staphylococcal protein A resulting in the activation of NF kappa B, AP-1, and downstream gene transcription. Exp Dermatol. 2011;20:48–52. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

