Traits of Bathy Phytochromes and Application to Bacterial Optogenetics
- PMID: 40645610
- PMCID: PMC12362586
- DOI: 10.1021/acssynbio.5c00337
Traits of Bathy Phytochromes and Application to Bacterial Optogenetics
Abstract
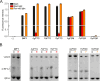
Phytochromes are photoreceptors sensitive to red and far-red light, found in a wide variety of organisms, including plants, fungi, and bacteria. Bacteriophytochromes (BphPs) can be switched between a red light-sensitive Pr state and a far-red light-sensitive Pfr state by illumination. In so-called prototypical BphPs, the Pr state functions as the thermally favored resting state, whereas Pfr is more stable in bathy BphPs. The prototypical DrBphP from Deinococcus radiodurans has been shown to be compatible with different output module types. Even though red light-regulated optogenetic tools are available, like the pREDusk system based on the DrBphP photosensory module, far-red light-modulated variants are still rare. Here, we study the underlying contributors to bathy over prototypical BphP behavior by way of various chimeric constructs between pREDusk and representative bathy BphPs. We pinpoint shared traits of the otherwise heterogeneous subgroup of bathy BphPs and highlight the importance of the sensor-effector linker in light modulation of histidine kinase activity. Informed by these data, we introduce the far-red light-activated system "pFREDusk", based on a histidine kinase activity governed by a bathy photosensory module. With this tool, we expand the optogenetic toolbox into wavelengths of increased sample and tissue penetration.
Keywords: histidine kinase; optogenetics; photoreceptor; phytochrome; protein chimera; two-component system.
Figures
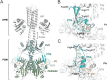




Similar articles
-
Engineered bacteriophytochrome heterodimers for research and applications.J Biol Chem. 2025 Aug;301(8):110452. doi: 10.1016/j.jbc.2025.110452. Epub 2025 Jul 4. J Biol Chem. 2025. PMID: 40617349 Free PMC article.
-
Cryo-EM structures of a bathy phytochrome histidine kinase reveal a unique light-dependent activation mechanism.Structure. 2024 Nov 7;32(11):1952-1962.e3. doi: 10.1016/j.str.2024.08.008. Epub 2024 Aug 30. Structure. 2024. PMID: 39216473
-
Darkness inhibits autokinase activity of bacterial bathy phytochromes.J Biol Chem. 2024 Apr;300(4):107148. doi: 10.1016/j.jbc.2024.107148. Epub 2024 Mar 9. J Biol Chem. 2024. PMID: 38462162 Free PMC article.
-
Aspects of Genetic Diversity, Host Specificity and Public Health Significance of Single-Celled Intestinal Parasites Commonly Observed in Humans and Mostly Referred to as 'Non-Pathogenic'.APMIS. 2025 Sep;133(9):e70036. doi: 10.1111/apm.70036. APMIS. 2025. PMID: 40923351 Free PMC article. Review.
-
Light therapies for acne.Cochrane Database Syst Rev. 2016 Sep 27;9(9):CD007917. doi: 10.1002/14651858.CD007917.pub2. Cochrane Database Syst Rev. 2016. PMID: 27670126 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

