Gut microbiota and brain-resident CD4+ T cells shape behavioral outcomes in autism spectrum disorder
- PMID: 40645945
- PMCID: PMC12254219
- DOI: 10.1038/s41467-025-61544-0
Gut microbiota and brain-resident CD4+ T cells shape behavioral outcomes in autism spectrum disorder
Abstract
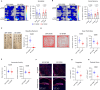
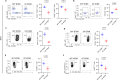
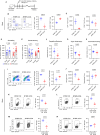
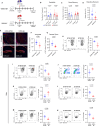
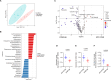
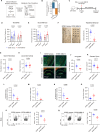
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by alterations in social, repetitive, and anxiety-like behaviors. While emerging evidence suggest a gut-brain etiology in ASD, the underlying mechanisms remain unclear. To dissect this axis, we developed a germ-free BTBR mouse model for ASD. The absence of gut microbiota in male mice ameliorates ASD-associated behaviors and reduces populations of inflammatory brain-resident T cells. Additionally, CD4+ T cell depletion mitigates neuroinflammation and ASD behaviors, suggesting a gut-immune-brain axis. We identify several microbial and metabolic regulators of ASD, particularly those relevant to the glutamate/GABA ratio and 3-hydroxyglutaric acid. Using an in silico metabolite prediction model, we propose Limosilactobacillus reuteri IMB015 (IMB015) to be a probiotic candidate. Administration of IMB015 reduces the glutamate/GABA ratio and neuroinflammation, resulting in improved behaviors. Here we report a gut-immune-brain axis in which the gut microbiota and its metabolites can modulate brain-resident immune cells and ASD-associated behaviors.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.L., S.L., and S.Y.C. are employees of ImmunoBiome. S.-H.I. is the founder and major shareholder of ImmunoBiome. The other authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

