TRF1 relies on fork reversal to prevent fragility at human telomeres
- PMID: 40645989
- PMCID: PMC12254382
- DOI: 10.1038/s41467-025-61828-5
TRF1 relies on fork reversal to prevent fragility at human telomeres
Abstract
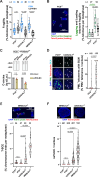
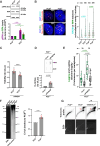
Telomeres pose challenges during replication, with converging forks unlikely to resolve issues. Depleting TRF1 results in fragile telomeres, yet its exact role in telomere replication remains unclear. In our cellular model, insufficient TRF1 density at long telomeres leads to telomere fragility that is alleviated by restoring telomeric TRF1 levels. Our findings indicate that TRF1 mitigates lagging strand telomere fragility through fork reversal in a process involving telomerase activity, rather than merely alleviating fork barriers. Additionally, TFIIH, a crucial partner of TRF1, aids in restarting replication on the leading strand after fork reversal. When fork reversal is compromised, PrimPol-mediated repriming rescues fragility at leading strand telomeres, revealing a new role for this enzyme in human telomere replication. Lastly, our findings indicate that the TRF1-mediated decrease in telomere fragility is dependent on RNA:DNA hybrids, likely facilitating fork restart.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- WWCR 24-0166/AICR_/Worldwide Cancer Research/United Kingdom
- K99-R00CA252447/U.S. Department of Health & Human Services | NIH | NCI | Division of Cancer Epidemiology and Genetics, National Cancer Institute (National Cancer Institute Division of Cancer Epidemiology and Genetics)
- R01 CA234047/CA/NCI NIH HHS/United States
- AIRC IG 28954/Associazione Angela Serra per la Ricerca sul Cancro (Associazione Angela Serra)
- AG0773424, CA234047, P30CA014195 and GM142173/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Research Materials

