Host albumin redirects Candida albicans metabolism to engage an alternative pathogenicity pathway
- PMID: 40646002
- PMCID: PMC12254302
- DOI: 10.1038/s41467-025-61701-5
Host albumin redirects Candida albicans metabolism to engage an alternative pathogenicity pathway
Abstract
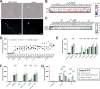
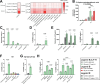
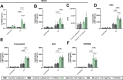
Pathogenicity mechanisms of the yeast Candida albicans involve filamentous growth, adhesion, invasion, and toxin production. Interestingly, clinical isolates, and other Candida spp., can cause infection independent of filamentation or toxin production. These strains and species often are characterized as avirulent ex vivo, yet this does not correlate with their potential to cause infection. We hypothesized that specific host factors, which trigger pathogenicity in vivo, are absent in in vitro infection models and thereby clinical isolates can seem avirulent ex vivo. We investigated how albumin, the most abundant protein in humans, impacts infection and cytotoxic potential of C. albicans in vitro. The presence of albumin induces otherwise non-damaging and non-filamentous clinical isolates to cause host cell cytotoxicity. Moreover, avirulent deletion mutants deficient in filamentation, adhesion, or toxin production are restored in their cytotoxicity by albumin. This involves transcriptional and metabolic reprogramming of C. albicans, increasing biofilm formation and production of the oxylipin 13-hydroxyoctadecadienoic acid, driving host cell cytotoxicity. Collectively, our study uncoveres a pathogenicity mechanism by which C. albicans causes epithelial cytotoxicity independent of its conventional virulence mechanisms. This alternative pathogenicity strategy helps to explain the avirulence of clinical isolates ex vivo, when they are separated from the host environment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Peroumal, D. et al. Virulence and pathogenicity of a Candida albicans mutant with reduced filamentation. Cell Microbiol21, e13103 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- project no. 434385622 / GR 5617/1-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- CRR/TRR124 FungiNet C1 210879364/Deutsche Forschungsgemeinschaft (German Research Foundation)
- CRR/TRR124 FungiNet C2 210879364/Deutsche Forschungsgemeinschaft (German Research Foundation)
- CRR/TRR124 FungiNet A7 210879364/Deutsche Forschungsgemeinschaft (German Research Foundation)
- Jena School of Microbial Communication PhD extension grant/Carl-Zeiss-Stiftung (Carl Zeiss Foundation)
- project number S006424N DeVEnIR/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- Exploration Grant/Boehringer Ingelheim Stiftung (Boehringer Ingelheim Foundation)
- 847507 (HDM-FUN)/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- BMBF 03Z22JN11/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

