Defining subgroups of pediatric nephrotic patients with urine proteomics
- PMID: 40646010
- PMCID: PMC12393002
- DOI: 10.1038/s41598-025-05150-6
Defining subgroups of pediatric nephrotic patients with urine proteomics
Abstract
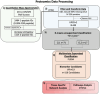
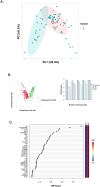
The molecular pathophysiology of nephrotic syndrome remains largely elusive in pediatric patients. While most children with minimal change disease (MCD) show favorable responses to immunosuppressive therapy, those with focal segmental glomerulosclerosis (FSGS) often exhibit poorer treatment responses, with many experiencing either partial remission or no remission of proteinuria. The need for reliable glomerular disease biomarkers to predict treatment response and understand molecular pathways governing responsiveness and resistance is a critical unmet need in pediatric nephrology. In this study, we sought to characterize urine proteomes in children with MCD and FSGS to identify biomarkers distinguishing disease activity and associated molecular pathways. Using quantitative proteomics, urine proteins from children with MCD and FSGS in the CureGN Study were identified and correlated with disease onset and activity. Unbiased cluster analyses of nephrotic urine proteomes demonstrated a cluster with relatively increased immune response and complement proteins, suggesting important distinctions in disease characteristics within the nephrotic subgroups. These analyses yielded patient subpopulations with proteinuria and distinct urine proteome differences associated with 116 proteins exerting cluster separation in the multivariate analyses. These findings highlight the potential of unsupervised clustering to identify disease subgroups and provide insights into the underlying molecular heterogeneity within nephrotic syndrome, paving the way for more tailored therapeutic strategies and improved patient management.
Keywords: FSGS; MCD; Nephrotic proteinuria; Pediatric glomerular disease; Proteomics; Urine biomarker.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

