A hybrid YOLO-UNet3D framework for automated protein particle annotation in Cryo-ET images
- PMID: 40646021
- PMCID: PMC12254289
- DOI: 10.1038/s41598-025-09522-w
A hybrid YOLO-UNet3D framework for automated protein particle annotation in Cryo-ET images
Abstract
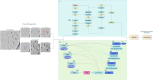
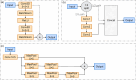
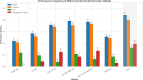
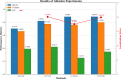
Accurate localization and identification of protein complexes in cryo-electron tomography (cryo-ET) volumes are essential for understanding cellular functions and disease mechanisms. However, automated annotation of these macromolecular assemblies remains challenging due to low signal-to-noise ratios, missing wedge artifacts, heterogeneous backgrounds, and structural diversity. In this study, we present a hybrid framework integrating You Only Look Once (YOLO) object detection with UNet3D volumetric segmentation, enhanced by density-based spatial clustering of applications with noise (DBSCAN) post-processing for automated protein particle annotation in cryo-ET volumes. Our approach combines YOLO's efficient region proposal capabilities with UNet3D's powerful 3D feature extraction through a dual-branch architecture featuring optimized Spatial Pyramid Pooling-Fast (SPPF) modules and asymmetric feature splitting. Extensive experiments on the Chan Zuckerberg Initiative Imaging (CZII) cryo-ET dataset demonstrate that our method significantly outperforms existing state-of-the-art approaches, including DeepFinder, standard UNet3D, YOLOv5-3D, and 3D ResNet models, achieving a mean recall of 0.8848 and F4-score of 0.7969. The framework demonstrates robust performance across various protein particle types and imaging conditions, offering a promising technical solution for high-throughput structural biology workflows requiring accurate macromolecular annotation in cellular cryo-ET data.
Keywords: Cryo-electron tomography; Deep learning; Density clustering; Protein particle detection; UNet3D; YOLO.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
An approach for coherent periodogram averaging of tilt-series data for improved contrast transfer function estimation.FEBS Open Bio. 2025 Aug;15(8):1209-1218. doi: 10.1002/2211-5463.70050. Epub 2025 May 8. FEBS Open Bio. 2025. PMID: 40341839 Free PMC article.
-
Beam image-shift accelerated data acquisition for near-atomic resolution single-particle cryo-electron tomography.Nat Commun. 2021 Mar 30;12(1):1957. doi: 10.1038/s41467-021-22251-8. Nat Commun. 2021. PMID: 33785757 Free PMC article.
-
CryoDRGN-AI: neural ab initio reconstruction of challenging cryo-EM and cryo-ET datasets.Nat Methods. 2025 Jul;22(7):1486-1494. doi: 10.1038/s41592-025-02720-4. Epub 2025 Jun 26. Nat Methods. 2025. PMID: 40571741
-
Cryo-electron tomography: Challenges and computational strategies for particle picking.Curr Opin Struct Biol. 2025 Aug;93:103113. doi: 10.1016/j.sbi.2025.103113. Epub 2025 Jul 9. Curr Opin Struct Biol. 2025. PMID: 40639056 Review.
-
A practical look at cryo-electron tomography image processing: Key considerations for new biological discoveries.Curr Opin Struct Biol. 2025 Aug;93:103116. doi: 10.1016/j.sbi.2025.103116. Epub 2025 Jul 8. Curr Opin Struct Biol. 2025. PMID: 40633126 Review.
References
-
- Rose, K. et al. In situ cryo-et visualization of mitochondrial depolarization and mitophagic engulfment. bioRxiv 2025–03 (2025)
-
- Kapnulin, L., Heimowitz, A. & Sharon, N. Outlier removal in cryo-em via radial profiles. J. Struct. Biol. 108172 (2025) - PubMed
MeSH terms
Substances
Grants and funding
- 20241501076/Zibo Medical and Health Research Project
- 2024KJJ044/the Youth Innovation Team Development Plan of Shandong
- H2023209049/the Natural Science Foundation of Hebei Province
- 24ZBSK091/the Zibo City Social Sciences Planning Project
- 2024355/the Scientific Research Project of Hebei Provincial Administration of Traditional Chinese Medicine
LinkOut - more resources
Full Text Sources

