Multomic analysis reveals the potential of LAG3 as a prognostic and immune biomarker and its validation in osteosarcoma
- PMID: 40646089
- PMCID: PMC12254381
- DOI: 10.1038/s41598-025-10290-w
Multomic analysis reveals the potential of LAG3 as a prognostic and immune biomarker and its validation in osteosarcoma
Erratum in
-
Correction: Multomic analysis reveals the potential of LAG3 as a prognostic and immune biomarker and its validation in osteosarcoma.Sci Rep. 2026 Jan 14;16(1):1753. doi: 10.1038/s41598-026-36146-5. Sci Rep. 2026. PMID: 41535504 Free PMC article. No abstract available.
Abstract
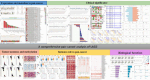
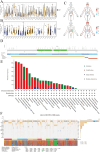
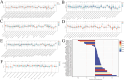
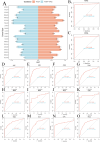
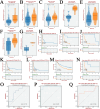
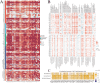
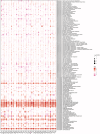
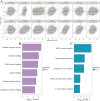
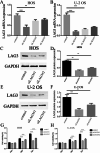
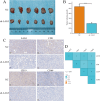
Lymphocyte activation gene 3 (LAG3) is a member of the immunoglobulin superfamily and has been implicated in the development, growth, and progression of several cancers. However, the biological role of LAG3 has not been investigated in pan-cancer datasets. We sought to perform a comprehensive bioinformatics analysis of pan-cancer datasets to determine the relationship of LAG3 with patient survival prognosis, tumor microenvironment, immunotherapy responsiveness, and mechanisms regulating tumorigenesis. We used TCGA, GTEx, TIMER2, GDSC, CTRP, and TISCH databases and online websites to extract data on LAG3 in a variety of cancers, and analyzed pan-cancer patient datasets to explore not only the correlation between LAG3 expression and clinical stage, diagnosis, and prognosis of cancers, but also the correlation between LAG3 expression, gene variants, methylation status, tumor stemness, and tumor immunity. The biological functions of LAG3 in osteosarcoma cells were determined by in vitro CCK-8, wound healing and transwell assays. Finally, through in vivo experiments, the study preliminarily explored the impact of LAG3 on osteosarcoma and its correlation with immune genes. Pan-cancer analysis showed that LAG3 expression was up-regulated in a variety of cancers, and the expression of LAG3 was closely related to the clinical stage, diagnosis and prognosis of cancers. GO and KEGG enrichment analyses showed that LAG3 was enriched in inflammatory, metabolic, and immune signaling pathways in a variety of cancers. Meanwhile, LAG3 expression not only has an impact on patient immunotherapy prognosis and immunotherapy response, but also has a significant effect on drug sensitivity. In vitro experiments have shown that LAG3 promotes the proliferation, migration and invasion of osteosarcoma cells. In vivo xenotransplantation experiments further confirmed that LAG3 promotes the growth of osteosarcoma, and the expression of LAG3 is positively correlated with CD8, CD19, and CD68. Our study suggests that LAG3 is a promising marker for cancer diagnosis, prognosis, and treatment.
Keywords: Diagnosis; Immune; LAG3; Pan-cancer; Prognosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
- 82002840/National Natural Science Foundation of China
- 82002300/National Natural Science Foundation of China
- 2024HNSLXRY08/Advanced Scientific Research Foundation for the Returned Overseas Chinese Scholars in Henan Province
- LHGJ20240046/Key Scientific and Technological Projects in Henan Province
- YXKC2021046/Henan Province Health Science and Technology innovation outstanding young talents training program
LinkOut - more resources
Full Text Sources
Medical
Research Materials

