A genome-wide association study identifies an African-specific locus on chromosome 21q22.12 associated with Burkitt lymphoma risk and survival
- PMID: 40646133
- PMCID: PMC12380610
- DOI: 10.1038/s41375-025-02690-8
A genome-wide association study identifies an African-specific locus on chromosome 21q22.12 associated with Burkitt lymphoma risk and survival
Abstract
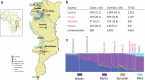
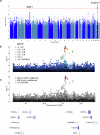
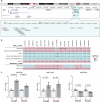
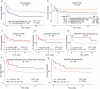
Burkitt lymphoma (BL) is a B-cell malignancy that disproportionately affects children in sub-Saharan Africa. We performed a genome-wide association study (GWAS) in a combined set of 800 childhood cases and 3865 controls in East Africa, controlling for age, sex, country, population-specific principal components, and a genetic relationship matrix. This analysis identified a BL-protective region within chromosome 21q22.12 tagged by the rs111457485-T allele (odds ratio [OR] = 0.57; p = 5.7 × 10-9). The results were robust in standard meta-analysis (OR = 0.57, p < 1.6 × 10-8), sensitivity analyses (removing genomic outliers and related individuals), and after adjustment for Epstein-Barr virus (EBV) status. Genomic analyses revealed long-range (over ~700 kb) chromatin interactions between the chr21q22.12 locus and the RUNX1-P1 promoter region. The African-specific rs2242780-C allele (r2 = 0.69 with the rs111457485-T allele in the study controls) showed increased enhancer activity in in-vitro Luciferase reporter assays (p = 4.5 × 10-10), nominating it as the likely functional variant for the BL-associated loci. In addition to the association with reduced BL risk in GWAS (OR = 0.62, p = 2.24 × 10-8), the rs2242780-C allele was also associated with better survival in patients with abdominal-only BL in exploratory analyses (hazard ratio = 0.39, p = 0.038, 106 patients, 59 deaths). Our GWAS uncovered novel BL-protective loci near RUNX1, offering insights into the genetic etiology of BL in African children.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
-
- López C, Burkhardt B, Chan JKC, Leoncini L, Mbulaiteye SM, Ogwang MD, et al. Burkitt lymphoma. Nat Rev Dis Prim. 2022;8:78. - PubMed
-
- Linet MS, Brown LM, Mbulaiteye SM, Check D, Ostroumova E, Landgren A, et al. International long-term trends and recent patterns in the incidence of leukemias and lymphomas among children and adolescents ages 0-19 years. Int J Cancer. 2016;138:1862–74. - PubMed
-
- Mbulaiteye SM, Devesa SS. Burkitt lymphoma incidence in five continents. Hemato. 2022;3:434–53.
-
- Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet. 1964;1:702–3. - PubMed
-
- Bornkamm GW. Epstein-Barr virus and the pathogenesis of Burkitt’s lymphoma: more questions than answers. Int J Cancer. 2009;124:1745–55. - PubMed
MeSH terms
Substances
Grants and funding
- HHSN261201100007I/CA/NCI NIH HHS/United States
- HHSN261201100063C/CA/NCI NIH HHS/United States
- SUpport to SJR/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- HHSN261201100063C, HHSN261201100007I, 75N910D00024/U.S. Department of Health & Human Services | NIH | NCI | Division of Cancer Epidemiology and Genetics, National Cancer Institute (National Cancer Institute Division of Cancer Epidemiology and Genetics)
- 75N910D00024/U.S. Department of Health & Human Services | NIH | NCI | Division of Cancer Epidemiology and Genetics, National Cancer Institute (National Cancer Institute Division of Cancer Epidemiology and Genetics)
LinkOut - more resources
Full Text Sources

