Comprehensive analysis of Mendelian randomization and scRNA-seq identify key prognostic genes and relevant functional roles in colorectal cancer
- PMID: 40646181
- PMCID: PMC12254384
- DOI: 10.1038/s41598-025-10354-x
Comprehensive analysis of Mendelian randomization and scRNA-seq identify key prognostic genes and relevant functional roles in colorectal cancer
Abstract
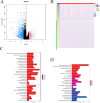
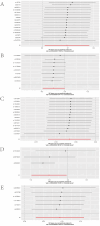
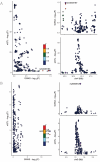
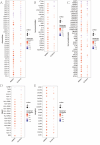
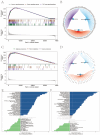
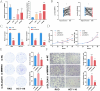
The prognosis of advanced CRC is poor, and identifying key genes related to CRC is vital for improving CRC prognosis. Our research used univariate Cox analysis and Mendelian randomization (MR) analysis to identify key prognostic genes in CRC. Multiple datasets such as the nomogram model and single-cell sequencing (scRNA-seq) were used to investigate the potential molecular mechanisms of the key genes. The expression levels were confirmed by using quantitative real-time polymerase chain reaction (qRT-PCR). The biological functions and effect on prognosis of the identified prognostic genes were also explored. MMRN1 and SLC6A19 were identified as key prognostic genes for CRC. Subsequently, the nomogram model demonstrated that MMRN1 and SLC6A19 can strongly predict survival. Further examination with multiple datasets elucidated the potential molecular mechanisms of the key prognostic genes, revealing a close association with immune cell infiltration. MMRN1 is enriched in classic CRC signaling pathways, whereas SLC6A19 is enriched in metabolism-related pathways. They are closely linked to immune cell infiltration levels and significantly influence the immune microenvironment in CRC. These key prognostic genes are significantly correlated with chemotherapeutic drug sensitivity and present promising opportunities for CRC therapy. The expression of both key genes was also observed in the scRNA-seq data of CRC. Finally, qRT-PCR validation revealed that MMRN1 is markedly downregulated and SLC6A19 is significantly upregulated in CRC. Lower expression of MMRN1 and higher expression of SLC6A19 significantly promoted the proliferation and metastasis of colorectal cancer cells. Our study identified MMRN1 and SLC6A19 as potential key prognostic genes for CRC, as they can reliably predict the prognosis of CRC. Furthermore, the potential molecular mechanisms of MMRN1 and SLC6A19 were revealed, suggesting new drug targets and therapeutic directions for managing prognosis.
Keywords: MMRN1; SLC6A19; Colorectal cancer; Immune microenvironment; Mendelian randomization; Prognosis; Single-cell RNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interest: The authors declare no competing interests. Ethical approval and consent to participate: The Ethics Committee of the First Affiliated Hospital of Ningbo University granted approval for this project (No.KY20220101). All human studies were carried out in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from the patients.
Figures




Similar articles
-
Construction of a New Ferroptosis-related Prognosis Model for Survival Prediction in Colorectal Cancer.Curr Med Chem. 2025;32(20):4132-4146. doi: 10.2174/0109298673296767240116215814. Curr Med Chem. 2025. PMID: 38362684
-
Potential of SPHK1 as a prognostic marker and therapeutic target in colorectal cancer: insights from bioinformatics and experimental analysis.Int J Surg. 2025 Jul 1;111(7):4550-4575. doi: 10.1097/JS9.0000000000002506. Epub 2025 May 28. Int J Surg. 2025. PMID: 40434736
-
Identification of biomarkers for Laryngeal squamous cell carcinoma through Mendelian randomization and integrated bioinformatics analysis.Discov Oncol. 2025 Jul 18;16(1):1364. doi: 10.1007/s12672-025-03114-w. Discov Oncol. 2025. PMID: 40679719 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

