Hypoxia- and lactate metabolism-associated prognostic and therapeutic signature in pancreatic cancer
- PMID: 40646423
- PMCID: PMC12254452
- DOI: 10.1007/s12672-025-02873-w
Hypoxia- and lactate metabolism-associated prognostic and therapeutic signature in pancreatic cancer
Abstract
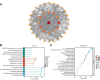
Background: Hypoxia and lactate metabolism products are critical components of the tumor microenvironment in pancreatic cancer (PC), influencing tumor invasiveness, metastasis, and treatment resistance. This study aims to explore the role of hypoxia- and lactate metabolism-related genes (HLRGs) in predicting overall survival and guiding treatment for PC patients.
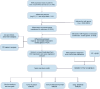
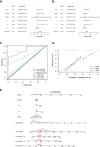
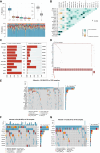
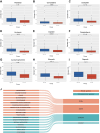
Methods: Gene expression and clinical data from PC patients were obtained from TCGA, ICGC, and GEO. Normal pancreatic tissue data were sourced from GTEx. Differential expression analysis was performed on the merged TCGA-PAAD and GTEx cohorts to identify differentially expressed genes (DEGs). We performed an intersection analysis between the DEGs and the HLRGs obtained from the MsigDB database to identify the DEGs associated with hypoxia and lactate metabolism in PC. A prognostic model was developed using random survival forests, Cox regression, and LASSO analysis in the TCGA-PAAD cohort. The model was externally validated in the ICGC-PACA and GSE85916 cohorts. Risk stratification was performed, and the differences between subgroups in tumor mutational burden, immune microenvironment, and drug response were analyzed. RT-qPCR validated the key genes expression differences.
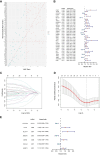
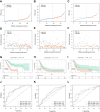
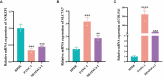
Results: A prognostic model based on HLRGs (SLC7A7, PYGL, HS3ST1, DDIT4, CYP27A1, ANKZF1, COL5A1) was established. High-risk patients exhibited worse prognosis, higher tumor mutational burden, and better response to anti-PD-L1 therapy, while low-risk patients exhibited higher immune infiltration and increased chemotherapy sensitivity. RT-qPCR confirmed that SLC7A7 and COL5A1 were upregulated, while ANKZF1 was downregulated in PC.
Conclusions: We developed an HLRGs-based prognostic model that predicts overall survival and guides treatment strategies, contributing to precision therapy in PC.
Keywords: Drug sensitivity; Hypoxia; Lactate metabolism; Pancreatic cancer; Prognosis; Tumor immune microenvironment; Tumor mutational burden.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study utilizes open-access databases and does not engage in original research involving human participants or animals. Competing interests: The authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
