Accurate determination of breed origin of alleles in a simulated smallholder crossbred dairy cattle population
- PMID: 40646444
- PMCID: PMC12247292
- DOI: 10.1186/s12711-025-00985-z
Accurate determination of breed origin of alleles in a simulated smallholder crossbred dairy cattle population
Abstract
Background: Accurate assignment of breed origin of alleles (BOA) at a heterozygote locus may help to introduce a resilient or adaptive haplotype in crossbreeding. In this study, we developed and tested a method to assign breed of origin for individual alleles in crossbred dairy cattle. After generations of mating within and between local breeds as well as the importation of exotic bulls, five rounds of selected crossbred cows were simulated to mimic a dairy breeding program in the low- and middle-income countries (LMICs). In each round of selection, the alleles of those crossbred animals were phased and assigned to their breed of origin (being either local or exotic).
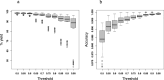
Results: Across all core lengths and modes of phasing (with offset-move 50% of the core length forward or no-offset), the average percentage of alleles correctly assigned a breed origin was 95.76%, with only 1.39% incorrectly assigned and 2.85% missing or unassigned. On consensus, the average percentage of alleles correctly assigned a breed origin was 93.21%, with only 0.46% incorrectly assigned and 6.33% missing or unassigned. This high proportion of alleles correctly assigned a breed origin resulted in a high core-based mean accuracy of 0.99 and a very high consensus-based (most frequently observed assignment across all the scenarios) mean accuracy of 1.00. The algorithm's assignment yield and accuracy were affected by the choice of threshold levels for the best match of assignments. The threshold level had the opposite effect on assignment yield and assignment accuracy. A less stringent threshold generated higher assignment yields and lower assignment accuracy.
Conclusions: We developed an algorithm that accurately assigns a breed origin to alleles of crossbred animals designed to represent breeding programs in the LMICs. The developed algorithm is straightforward in its application and does not require prior knowledge of pedigree, which makes it more relevant and applicable in LMICs breeding programs.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: RCG and JMH are now employed by Bayer Crop Science.
Figures





References
-
- Leroy G, Baumung R, Boettcher P, Scherf B, Hoffmann I. Review: sustainability of crossbreeding in developing countries; definitely not like crossing a meadow. Animal. 2016;10:262–73. - PubMed
-
- VanRaden PM, Cooper TA. Genomic evaluations and breed composition for crossbred US dairy cattle. Interbull Bull. 2015;49:19–23.
-
- Kuehn LA, Keele JW, Bennett GL, McDaneld TG, Smith TPL, Snelling WM, et al. Predicting breed composition using breed frequencies of 50,000 markers from the US meat animal research center 2,000 bull project. J Anim Sci. 2011;89:1742–50. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

