Genome-Wide Association Study for Weight-Related Traits in Scylla paramamosain Using Whole-Genome Resequencing
- PMID: 40646730
- PMCID: PMC12248745
- DOI: 10.3390/ani15131829
Genome-Wide Association Study for Weight-Related Traits in Scylla paramamosain Using Whole-Genome Resequencing
Abstract
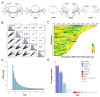
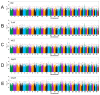
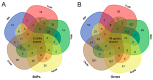
Weight traits serve as key economic indicators for assessing growth performance and commercial quality in the mud crab Scylla paramamosain, yet the genetic basis of these traits remains poorly characterized. Here, we performed whole-genome resequencing on 323 individuals and conducted genome-wide association studies (GWAS) on five weight-related traits: (1) body-related traits, including body weight (BW), trunk weight (TruW), and weight excluding chelae (WEC); (2) appendage-related traits, containing appendage weight (AppW) and cheliped weight (CheW). Significantly associated SNPs were primarily enriched on chromosomes 15, 22, 25, and 36. For body-related traits, we identified 45 shared candidate SNPs and 175 common candidate genes; appendage-related traits revealed 71 shared candidate SNPs, and 229 common genes were identified; and across all five traits, there were 9 shared candidate SNPs and 49 common genes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses indicated that shared functional terms/pathways among the five traits were mainly related to metabolism, development, and immunity. Body-related traits exhibited more unique GO terms and KEGG pathways associated with metabolism and immunity, whereas appendage-related traits showed some unique GO terms and KEGG pathways involved in development and morphogenesis. Among the candidate genes, we identified multiple genes associated with growth and development, metabolism, and immune responses. For example, the CCHa1R gene, common to carapace-related traits, is linked to feeding; the DCX-EMA gene, which is common to appendage-related traits, is connected to movement, and the MSTO1 gene is pertinent to muscle development. Among the candidate genes shared by all five traits, there are a series of genes concerning growth and development (such as NVD, CYP307A1, FGF1, NF2, ANKRD52) and immune responses (RGS10). These findings advance our understanding of the genetic architecture underlying decapod crustacean growth and provide valuable insights for optimizing sustainable breeding strategies in S. paramamosain.
Keywords: GWAS; Scylla paramamosain; weight-related traits; whole-genome resequencing.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

Similar articles
-
Heritability estimates and genome-wide association study of methane emission traits in Nellore cattle.J Anim Sci. 2024 Jan 3;102:skae182. doi: 10.1093/jas/skae182. J Anim Sci. 2024. PMID: 38967061 Free PMC article.
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Identification of new candidate genes affecting drip loss in pigs based on genomics and transcriptomics data.J Anim Sci. 2025 Jan 4;103:skaf177. doi: 10.1093/jas/skaf177. J Anim Sci. 2025. PMID: 40485044 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Zhao M., Wang W., Zhang F., Ma C., Liu Z., Yang M.H., Chen W., Li Q., Cui M., Jiang K., et al. A chromosome-level genome of the mud crab (Scylla paramamosain estampador) provides insights into the evolution of chemical and light perception in this crustacean. Mol. Ecol. Resour. 2021;21:1299–1317. doi: 10.1111/1755-0998.13332. - DOI - PubMed
-
- Keenan C., Davie P.J.F., Mann D.L. A revision of the genus Scylla de Haan, 1833 (Crustacea: Decapoda: Brachyura: Portunidae) Raffles Bull. Zool. 1998;46:217–245.
-
- Lovatelli A., Shelley C., Tobias-Quinitio E., Khor W., Chan D. Status, technological innovations, and industry development needs of mud crab (Scylla spp.) aquaculture; Proceedings of the FAO Expert Workshop; Singapore. 27–30 November 2023; Rome, Italy: FAO; 2023. FAO Fisheries and Aquaculture Proceedings, No. 73.
-
- Li Y., Ai C., Liu L. Aquaculture in China: Success Stories and Modern Trends. Wiley; Hoboken, NJ, USA: 2018. Mud crab, Scylla paramamosain China’s leading maricultured crab; pp. 226–233.
-
- FAO . Fishery and Aquaculture Statistics—Yearbook 2021. FAO; Rome, Italy: 2024. FAO Yearbook of Fishery and Aquaculture Statistics. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

