Integrated Microbiome-Metabolome Analysis Reveals Intestine-Liver Metabolic Associations in the Moustache Toad
- PMID: 40646872
- PMCID: PMC12248546
- DOI: 10.3390/ani15131973
Integrated Microbiome-Metabolome Analysis Reveals Intestine-Liver Metabolic Associations in the Moustache Toad
Abstract
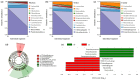
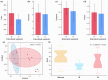
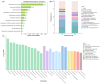
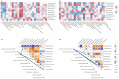
The intestinal microbiota regulates host metabolic homeostasis through production of bioactive microbial metabolites. These microorganisms facilitate digestion, enhance immune function, maintain osmoregulation, and support physiological balance via these bioactive compounds, thereby enhancing environmental adaptation. Our study investigated intestinal microbiota-liver metabolic interactions in Leptobrachium liui using 16S rRNA gene sequencing and non-targeted liquid chromatography-tandem mass spectrometry metabolomics. Key findings include (1) comparable alpha diversity but distinct microbial community structures between the small intestine (SI) and large intestine (LI), with the SI dominated by Enterobacteriaceae (72.14%) and the LI by Chitinophagaceae (55.16%); (2) segment-specific microbe-metabolite correlations, with predominantly positive correlations in the SI and complex patterns in the LI involving fatty acids, amino acids, and energy metabolites; and (3) significant correlations between specific bacterial families (Aeromonadaceae, Enterobacteriaceae, Chitinophagaceae) and hepatic metabolites related to fatty acid metabolism, amino acid synthesis, and energy pathways, indicating potential gut-liver axis associations. These findings provide insights into amphibian intestinal microbiota-hepatic metabolite associations and may inform future studies of host-microbe interactions.
Keywords: Leptobrachium liui; amphibian; gut–liver axis; intestinal microbiota; microbiome–metabolome.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- Kelly C.J., Zheng L., Campbell E.L., Saeedi B., Scholz C.C., Bayless A.J., Wilson K.E., Glover L.E., Kominsky D.J., Magnuson A. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

