Integrated Transcriptomic and Proteomic Analysis Reveals Differential Gene and Protein Expression and Signaling Pathways During a 20 Km Endurance Exercise and Recovery in Mongolian Horses
- PMID: 40646880
- PMCID: PMC12248615
- DOI: 10.3390/ani15131981
Integrated Transcriptomic and Proteomic Analysis Reveals Differential Gene and Protein Expression and Signaling Pathways During a 20 Km Endurance Exercise and Recovery in Mongolian Horses
Abstract
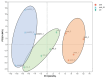
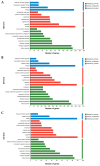
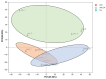
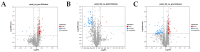
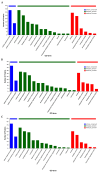
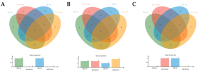
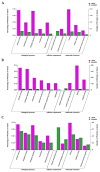
Mongolian horses are renowned for their remarkable endurance and ability to adapt to harsh environments. To delve deeper into the molecular mechanisms that underlie these traits, researchers conducted a comprehensive analysis of transcriptomic and proteomic changes in Mongolian horses at three distinct time points: before, immediately after, and 24 h following a 20 km run. The transcriptomic analysis uncovered significant variations in gene expression patterns across these time points. Specifically, 291 differentially expressed genes (DEGs) were identified when comparing pre-exercise to post-exercise conditions, 832 DEGs in the comparison between post-exercise and 24 h post-exercise, and 127 DEGs in the comparison of pre-exercise to 24 h post-exercise. Notably, key genes involved in metabolic activities and cellular proliferation, such as PI3K and LDHA, exhibited significant upregulation immediately after exercise but demonstrated a downward trend 24 h post-exercise. Concurrently, the proteomic analysis revealed 49 differentially expressed proteins (DEPs) in the pre-exercise versus post-exercise comparison, 61 DEPs in the post-exercise versus 24 h post-exercise comparison, and 101 DEPs in the pre-exercise versus 24 h post-exercise comparison. Some proteins, like PDK4 and GLUL, remained upregulated at 24 h post-exercise, whereas others, such as PFKM and LDHA, showed signs of recovery or downregulation. By integrating the transcriptomic and proteomic data, we were able to pinpoint overlapping DEGs/DEPs and implicate crucial signaling pathways, including the HIF-1 signaling pathway and glycolysis, in the molecular response of Mongolian horses to exercise. These findings offer insights into the endurance adaptation mechanisms of the Mongolian horse.
Keywords: Mongolian horses; adaptive response; endurance; exercise; proteomic analysis; transcriptomic analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Liu Y., Sinjab A., Min J., Han G., Paradiso F., Zhang Y., Wang R., Pei G., Dai Y., Liu Y., et al. Conserved spatial subtypes and cellular neighborhoods of cancer-associated fibroblasts revealed by single-cell spatial multi-omics. Cancer Cell. 2025;43:905–924. doi: 10.1016/j.ccell.2025.03.004. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

