Immunosuppressive Tumor Microenvironment of Osteosarcoma
- PMID: 40647416
- PMCID: PMC12248827
- DOI: 10.3390/cancers17132117
Immunosuppressive Tumor Microenvironment of Osteosarcoma
Abstract
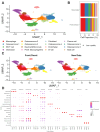
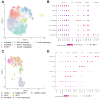
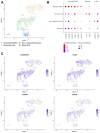
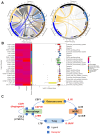
Background/Objectives: Osteosarcoma is the most common malignant bone tumor in children, characterized by a high degree of genomic instability, resulting in copy number alterations and genomic rearrangements without disease-defining recurrent mutations. Clinical trials based on molecular characterization have failed to find new effective therapies or improve outcomes over the last 40 years. Methods: To better understand the immune microenvironment of osteosarcoma, we performed single-cell RNA sequencing on six tumor biopsy samples, combined with a previously published cohort of six samples. Additional osteosarcoma samples were profiled using spatial transcriptomics for the validation of discovered subtypes and to add spatial context. Results: Analysis revealed immunosuppressive cells, including myeloid-derived suppressor cells (MDSCs), regulatory and exhausted T cells, and LAMP3+ dendritic cells. Conclusions: Using cell-cell communication modeling, we identified robust interactions between MDSCs and other cells, leading to NF-κB upregulation and an immunosuppressive microenvironment, as well as interactions involving regulatory T cells and osteosarcoma cells that promoted tumor progression and a proangiogenic niche.
Keywords: immune microenvironment; osteosarcoma; single-cell sequencing; spatial transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- D’Angelo S.P., Mahoney M.R., Van Tine B.A., Atkins J., Milhem M.M., Jahagirdar B.N., Antonescu C.R., Horvath E., Tap W.D., Schwartz G.K., et al. Nivolumab with or without Ipilimumab Treatment for Metastatic Sarcoma (Alliance A091401): Two Open-Label, Non-Comparative, Randomised, Phase 2 Trials. Lancet Oncol. 2018;19:416–426. doi: 10.1016/S1470-2045(18)30006-8. - DOI - PMC - PubMed
Grants and funding
- NA/CapoStrong Fund
- 19/18670-9/FAPESP
- P30 CA034196/CA/NCI NIH HHS/United States
- NA/The Jackson Laboratory CATch Program
- NA/Strawbridge Cancer Therapeutic Fund
- NA/T. T. & W.F. Chao Foundation
- NA/John S Dunn Research Foundation
- P30 CA06934/NH/NIH HHS/United States
- RRID:SCR_022035/Flow Cytometry shared resources
- RRID:SCR_021984/University of Colorado Bioinformatics and Biostatistics Shared Resource
- U01CA253553/NH/NIH HHS/United States
- NA/St. Baldrick's Foundation Scholar Award
- NA/National Pediatric Cancer Foundation
LinkOut - more resources
Full Text Sources

