Oncogenic Activity and Sorafenib Sensitivity of ARAF p.S214C Mutation in Lung Cancer
- PMID: 40647542
- PMCID: PMC12248905
- DOI: 10.3390/cancers17132246
Oncogenic Activity and Sorafenib Sensitivity of ARAF p.S214C Mutation in Lung Cancer
Abstract
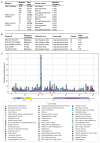
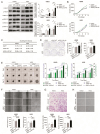
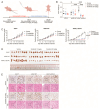
Background/Objectives: RAF pathway aberrations are one of the hallmarks of lung cancer. Sorafenib is a multi-kinase inhibitor targeting the RAF pathway and is FDA-approved for several cancers, yet its efficacy in lung cancer is controversial. Previous clinical research showed that a ARAF p.S214C mutation exhibited exceptional responsiveness to sorafenib in lung adenocarcinoma. Methods: Considering this promising clinical potential, the oncogenic potential and sorafenib response of the ARAF p.S214C mutation were investigated using lung cancer models. ARAF p.S214C mutant, ARAF wild-type (WT), and EGFP control genes were ectopically expressed in lung adenocarcinoma cell lines retroviral transduction. In vitro and in vivo sorafenib sensitivity studies were performed, followed by transcriptomics and proteomics analyses. Results: Compared to the ARAF-WT and EGFP-engineered cells, the ARAF p.S214C-engineered cells activated Raf-MEK-ERK signaling and exhibited enhanced oncogenic potential in terms of in vitro cell proliferation, colony and spheroid formation, migration, and invasion abilities, as well as in vivo tumorigenicity. The ARAF p.S214C-engineered cells also displayed heightened sensitivity to sorafenib in vitro and in vivo. RNA sequencing and reverse-phase protein array analyses demonstrated elevated expression of genes and proteins associated with tumor aggressiveness in the ARAF p.S214C mutants, and its sorafenib sensitivity was likely moderated through inhibition of the cell cycle and DNA replication. The ERK and PI3K signaling pathways were also significantly deregulated in the ARAF p.S214C mutants regardless of sorafenib treatment. Conclusions: This study demonstrates the oncogenicity and sorafenib sensitivity of the ARAF p.S214C mutation in lung cancer cells, which may serve as a biomarker for predicting the sorafenib response in lung cancer patients. Importantly, investigating the gene-drug sensitivity pairs in clinically exceptional responders may guide and accelerate personalized cancer therapies based on specific tumor mutations.
Keywords: ARAF p.S214C mutation; lung cancer; oncogenicity; sorafenib sensitivity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

