Overexpression of SEPALLATA3-like Gene SnMADS37 Generates Green Petal-Tip Flowers in Solanum nigrum
- PMID: 40647900
- PMCID: PMC12251700
- DOI: 10.3390/plants14131891
Overexpression of SEPALLATA3-like Gene SnMADS37 Generates Green Petal-Tip Flowers in Solanum nigrum
Abstract
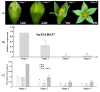
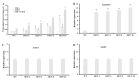
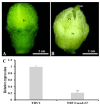
The SEPALLATA3 (SEP3)-like MADS-box genes play crucial roles in determining petal identity and development in the petunia and tomato of Solanaceae. Solanum nigrum is a self-pollinating plant in the Solanaceae family, and produces white flowers. However, the mechanisms controlling the transition from green to white petals during flower development remain poorly understood. In this study, we isolated a flower-specific SEP3-like gene, SnMADS37, from S. nigrum, and investigated its potential role in chlorophyll metabolism during petal development. Our results show that quantitative RT-PCR analysis demonstrates that SnMADS37 is exclusively expressed in petals and stamens during early floral bud development. Overexpression of SnMADS37 clearly enhanced the number of petals, promoting the formation of additional petal-like tissues in stamens and extra organs in some fruits. Moreover, fully opened transformed petals exhibited notable chlorophyll accumulation at their tips and veins, whereas silencing of Snmads37 clearly inhibited petal expansion and reduced green pigmentation in early flower buds. Additionally, the transformed green petals exhibited distinct conical epidermal cells in the green regions, similar to wild type (WT) petals. Our results demonstrate that SnMADS37 plays a critical role in regulating petal identity, expansion, and chlorophyll metabolism during petal development. These findings provide new insights into the functional diversification of SEP3-like MADS-box genes in angiosperms.
Keywords: MADS-box; SEPALLATA3-like gene SnMADS37; Solanaceae; Solanum nigrum; chlorophyll accumulation; green petal-tip flower; overexpression; petal development.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Edmonds J., Chweya J.A. Promoting the Conservation and Use of Underutilized and Neglected Crops. 15th ed. Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute; Rome, Italy: 1997. Black nightshades. Solanum nigrum L. and related species.
-
- Lehmann C., Biela C., Töpfl S., Jansen G., Vögel R. Solanum scabrum a potential source of a coloring plant extract. Euphytica. 2007;158:189–199. doi: 10.1007/s10681-007-9442-2. - DOI
-
- Jabamalairaj A., Priatama R.A., Heo J., Park S.J. Medicinal metabolites with common biosynthetic pathways in Solanum nigrum. Plant Biotech. Rep. 2019;13:315–327. doi: 10.1007/s11816-019-00549-w. - DOI
-
- Sommer H., Beltrán J.P., Huijser P., Pape H., Lönnig W.E., Saedler H., Schwarz-Sommer Z. Defciens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus, the protein shows homology to transcription factors. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

