Genome-Wide Identification of the DOG1 Gene Family in Pepper (Capsicum annuum) and Its Expression Profiles During Seed Germination
- PMID: 40647924
- PMCID: PMC12251688
- DOI: 10.3390/plants14131913
Genome-Wide Identification of the DOG1 Gene Family in Pepper (Capsicum annuum) and Its Expression Profiles During Seed Germination
Abstract
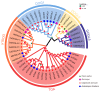
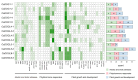
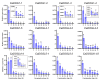
The DOG1 (Delay of Germination1) family plays key regulatory roles in seed dormancy and germination. However, a genome-wide analysis of DOG1 genes has not been performed for pepper (Capsicum annuum), one of the agriculturally important species, and no studies have been conducted to characterize their expression profiles. Based on C. annuum genome information, the identification and expression analysis of CaDOG1 gene family members through bioinformatics approaches can provide a theoretical foundation for subsequent studies on the biological functions of CaDOG1s and the improvement of seed traits in C. annuum breeding. In this study, a total of 13 CaDOG1 genes were identified in the C. annuum genome. Phylogenetic analysis showed that these CaDOG1s, along with DOG1s from thale cress (Arabidopsis thaliana), rice (Oryza sativa), and maize (Zea mays), were classified into four subgroups. All CaDOG1 genes were unevenly distributed on six C. annuum chromosomes, and they had relatively conserved exon-intron patterns, most with zero to one intron. According to the chromosomal distribution patterns and synteny analysis of the CaDOG1 genes, the CaDOG1 family expanded mainly through replication, which occurred predominantly after the divergence of dicotyledons and monocotyledons. Conserved motif analysis indicated that all encoded proteins contained Motif 2 and Motif 6, except for CaDOG1-3. Expression profile analysis using transcriptome data revealed that CaDOG1 genes were differentially expressed across various tissues and developmental stages, with notable involvement in flowers and seeds. Quantitative real-time PCR also revealed that all CaDOG1 genes were downregulated during seed germination, indicating that CaDOG1s may play negative roles in seed germination. Moreover, upon abscisic acid treatment, six CaDOG1 genes exhibited upregulation, while in response to ethylene, four CaDOG1 genes exhibited downregulation. Taken together, these findings provide an extensive description of the C. annuum DOG1 gene family and might facilitate further studies for elucidating their functions in seed germination.
Keywords: DOG1; bioinformatics; expression profiling; pepper (Capsicum annuum); seed germination.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
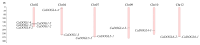




Similar articles
-
Genome-Wide Identification and Analysis of the AHL Gene Family in Pepper (Capsicum annuum L.).Int J Mol Sci. 2025 Jul 7;26(13):6527. doi: 10.3390/ijms26136527. Int J Mol Sci. 2025. PMID: 40650302 Free PMC article.
-
Comparative transcriptome analysis reveals the potential mechanism of seed germination promoted by trametenolic acid in Gastrodia elata Blume.Sci Rep. 2025 Jul 24;15(1):26869. doi: 10.1038/s41598-025-12269-z. Sci Rep. 2025. PMID: 40702084 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Comparative analysis of POD gene family and expression differentiation under NaCl, H2O2 and PEG stresses in sorghum.BMC Genomics. 2025 Jul 17;26(1):674. doi: 10.1186/s12864-025-11858-6. BMC Genomics. 2025. PMID: 40676507 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Lamont B.B., Pausas J.G. Seed dormancy revisited: Dormancy-release pathways and environmental interactions. Funct. Ecol. 2023;37:1106–1125. doi: 10.1111/1365-2435.14269. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

