Time-Series Metabolome and Transcriptome Analyses Reveal the Genetic Basis of Vanillin Biosynthesis in Vanilla
- PMID: 40647930
- PMCID: PMC12252122
- DOI: 10.3390/plants14131922
Time-Series Metabolome and Transcriptome Analyses Reveal the Genetic Basis of Vanillin Biosynthesis in Vanilla
Abstract
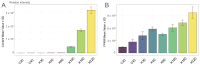
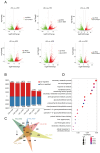
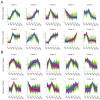
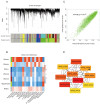
Vanillin, the principal aromatic compound in vanilla, is primarily derived from mature pods of vanilla (Vanilla planifolia Andrews). Although the biosynthetic pathway of vanillin has been progressively elucidated, the specific key enzymes and transcription factors (TFs) governing vanillin biosynthesis require further comprehensive investigation via combining transcriptomic and metabolomic analysis. For this study, V. planifolia (higher vanillin producer) and V. imperialis (lower vanillin producer) were selected. Time-series metabolomics analysis revealed 160-220 days after pollination (DAPs) as the critical phase for vanillin biosynthesis. Combined time-series transcriptome analysis revealed 984 upregulated differentially expressed genes (DEGs) in key periods, 2058 genes with temporal expression, and 4326 module genes through weighted gene co-expression network analysis (WGCNA), revealing six major classes of TFs: No Apical Meristem (NAC), Myb, WRKY, FLOWERING PROMOTING FACTOR 1-like (FPFL), DOF, and PLATZ. These TFs display strong regulatory relationships with the expression of key enzymatic genes, including P450s, COMT, and 4CL. The NAC TF family emerged as central regulators in this network, with NAC-2 (HPP92_014056) and NAC-3 (HPP92_012558) identified as key hub genes within the vanillin biosynthetic gene co-expression network. The findings of this study provide a theoretical foundation and potential target genes for enhancing vanillin production through genetic and metabolic engineering approaches, offering new opportunities for sustainable development in the vanilla industry and related applications.
Keywords: Vanilla planifolia Andrews; combined analysis; transcriptome and metabolome; vanillin.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Quezada-Euan J.J.G., Guerrero-Herrera R.O., Gonzalez-Ramirez R.M., MacFarlane D.W. Frequency and behavior of Melipona stingless bees and orchid bees (Hymenoptera: Apidae) in relation to floral characteristics of vanilla in the Yucatan region of Mexico. PLoS ONE. 2024;19:e0306808. doi: 10.1371/journal.pone.0306808. - DOI - PMC - PubMed
-
- Adame-Soto P.J., Arechiga-Carvajal E.T., Gonzalez-Herrera S.M., Moreno-Jimenez M.R., Rutiaga-Quinones O.M. Characterization of mating type on aroma production and metabolic properties wild Kluyveromyces marxianus yeasts. World J. Microbiol. Biotechnol. 2023;39:216. doi: 10.1007/s11274-023-03659-4. - DOI - PubMed
-
- da Silva Oliveira J.P., Garrett R., Koblitz M.G.B., Macedo A.F. Integrated targeted and untargeted metabolomics profiling of Vanilla species from the Atlantic Forest: Unveiling the bioeconomic potential of Vanilla cribbiana. Food Chem. 2025;464:141650. doi: 10.1016/j.foodchem.2024.141650. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

