Genome-Wide Identification and Expression Analysis of Aspartic proteases in Populus euphratica Reveals Candidates Involved in Salt Tolerance
- PMID: 40647939
- PMCID: PMC12252427
- DOI: 10.3390/plants14131930
Genome-Wide Identification and Expression Analysis of Aspartic proteases in Populus euphratica Reveals Candidates Involved in Salt Tolerance
Abstract
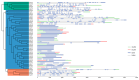
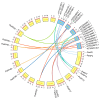
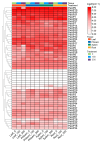
Aspartic proteases (APs) are among the four primary families of proteolytic enzymes found in plants, and they are essential for both stress response mechanisms and developmental activities. While the AP gene family has been studied in model plants like Arabidopsis, its characterization in woody species-particularly in extremophytes like Populus euphratica, remains limited. Moreover, the potential involvement of APs in salt tolerance mechanisms in trees is yet to be explored. In this research, 55 PeAPs were discovered and categorized into three distinct classes based on their conserved protein structures. The phylogenetic analysis revealed potential functions of AP genes derived from Arabidopsis thaliana, V. vinifera, and P. euphratica. Our findings indicate that PeAP possesses a well-conserved evolutionary background and contains numerous highly variable regions, making it an excellent candidate for the identification and systematic examination of woody trees. Additionally, motifs frequently found in aspartic proteases within the genome of P. euphratica may be linked to functional PeAPs. It appears that PeAPs are associated with specific gene functions. These genes are influenced by cis-elements, which may play a role in their responsiveness to phytohormone, stress adaptation maybe changed to these genes are regulated by cis-elements that may mediate their responsiveness to phytohormones, abiotic stress, and developmental cues. Our research offers the initial comprehensive analysis of the AP family in P. euphratica, emphasizing its potential functions in adapting to salt conditions. The findings uncover candidate PeAPs for genetic engineering to enhance salinity tolerance in woody crops.
Keywords: Aspartic proteases; Populus euphratica; gene family; salinity tolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Chen S., Hawighorst P., Sun J., Polle A. Salt tolerance in Populus: Significance of stress signaling networks, mycorrhization, and soil amendments for cellular and whole-plant nutrition. Environ. Exp. Bot. 2014;107:113–124. doi: 10.1016/j.envexpbot.2014.06.001. - DOI
-
- Watanabe S., Kojima K., Ide Y., Sasaki S. Effects of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro. Plant Cell Tissue Organ Cult. (PCTOC) 2000;63:199–206. doi: 10.1023/A:1010619503680. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

