Construction of an Overexpression Library for Chinese Cabbage Orphan Genes in Arabidopsis and Functional Analysis of BOLTING RESISTANCE 4-Mediated Flowering Delay
- PMID: 40647955
- PMCID: PMC12252378
- DOI: 10.3390/plants14131947
Construction of an Overexpression Library for Chinese Cabbage Orphan Genes in Arabidopsis and Functional Analysis of BOLTING RESISTANCE 4-Mediated Flowering Delay
Abstract
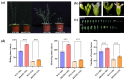
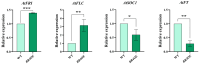
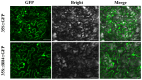
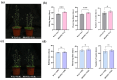
Orphan genes (OGs), which are unique to a specific taxon and have no detectable sequence homology to any known genes across other species, play a pivotal role in governing species-specific phenotypic traits and adaptive evolution. In this study, 20 OGs of Chinese cabbage (Brassica rapa OGs, BrOGs) were transferred into Arabidopsis thaliana by genetic transformation to construct an overexpression library in which 50% of the transgenic lines had a delayed flowering phenotype, 15% had an early flowering phenotype, and 35% showed no difference in flowering time compared to control plants. There were many other phenotypes attached to these transgenic lines, such as leaf color, number of rosette leaves, and silique length. To understand the impact of BrOGs on delayed flowering, BrOG142OE, which showed the most significantly delayed flowering phenotype, was chosen for further analysis, and BrOG142 was renamed BOLTING RESISTANCE 4 (BR4). In BR4OE, the expression of key flowering genes, including AtFT and AtSOC1, significantly decreased, and AtFLC and AtFRI expression increased. GUS staining revealed BR4 promoter activity mainly in the roots, flower buds and leaves. qRT-PCR showed that BR4 primarily functions in the flowers, flower buds, and leaves of Chinese cabbage. BR4 is a protein localized in the nucleus, cytoplasm, and cell membrane. The accelerated flowering time phenotype of BR4OE was observed under gibberellin and vernalization treatments, indicating that BR4 regulates flowering time in response to these treatments. These results provide a foundation for elucidating the mechanism by which OGs regulate delayed flowering and have significance for the further screening of bolting-resistant Chinese cabbage varieties.
Keywords: Arabidopsis; Chinese cabbage; delayed flowering; orphan genes; overexpression library.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources

