Strategies Used for the Discovery of New Microbial Metabolites with Antibiotic Activity
- PMID: 40649382
- PMCID: PMC12251386
- DOI: 10.3390/molecules30132868
Strategies Used for the Discovery of New Microbial Metabolites with Antibiotic Activity
Abstract
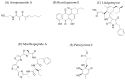
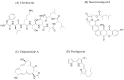
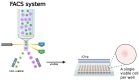
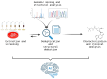
The discovery of new microbial metabolites is essential to combat the alarming rise in antimicrobial resistance and to meet emerging medical needs. This work critically reviews current strategies for identifying antimicrobial compounds, emphasizing the potential of microorganisms as a rich source of bioactive secondary metabolites. This review explores innovative methods, such as investigating extreme environments where adverse conditions favor the emergence of unique metabolites; developing techniques, like the iChip, to cultivate previously uncultivable bacteria; using metagenomics to analyze complex samples that are difficult to isolate; and integrates artificial intelligence to accelerate genomic mining, structural prediction, and drug discovery optimization processes. The importance of overcoming current challenges, such as replicating findings, low research investment, and the lack of adapted collection technologies, is also emphasized. Additionally, this work analyzes the crucial role of bacterial resistance and the necessity of a holistic approach involving new technologies, sustained investment, and interdisciplinary collaboration. This work emphasizes not only the current state of metabolite discovery but also the challenges that must be addressed to ensure a continuous flow of new therapeutic molecules in the coming decades.
Keywords: antibiotic; antimicrobial resistance; artificial intelligence; drug discovery; extremophiles; isolation Chip; metagenomics; microorganisms; natural products; secondary metabolites.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Székács A. Herbicides. Elsevier; Amsterdam, The Netherlands: 2021. Herbicide Mode of Action; pp. 41–86.
-
- Burg R.W., Miller B.M., Baker E.E., Birnbaum J., Currie S.A., Hartman R., Kong Y.-L., Monaghan R.L., Olson G., Putter I., et al. Avermectins, New Family of Potent Anthelmintic Agents: Producing Organism and Fermentation. Antimicrob. Agents Chemother. 1979;15:361–367. doi: 10.1128/AAC.15.3.361. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

