Transcriptomic Profiling of Zebrafish Mutant for cdkl5 Reveals Dysregulated Gene Expression Associated with Neuronal, Muscle, Visual and Skeletal Development
- PMID: 40649845
- PMCID: PMC12250029
- DOI: 10.3390/ijms26136069
Transcriptomic Profiling of Zebrafish Mutant for cdkl5 Reveals Dysregulated Gene Expression Associated with Neuronal, Muscle, Visual and Skeletal Development
Abstract
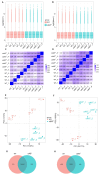
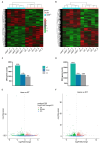
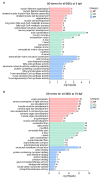
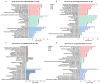
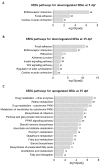
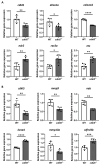
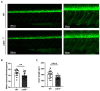
Zebrafish is a well-recognized model for studying human genetic disorders. Recently, we proposed the homozygous cdkl5sa21938 mutant zebrafish as a model of CDKL5 deficiency disorder (CDD), a developmental epileptic encephalopathy with diverse symptoms. This study aimed to explore Cdkl5-associated molecular mechanisms in zebrafish and assess their similarity to those in mammals. We conducted RNA sequencing on whole cdkl5-/- zebrafish and wild-type siblings at 5 and 35 days post-fertilization (dpf) to compare their gene expression profiles. Most significant differentially expressed genes (DEGs) were related to muscle, neuronal, and visual systems which are affected in CDD. Gene Ontology analysis revealed downregulated DEGs enriched in muscle development, extracellular matrix, and actin cytoskeleton functions at both stages, while upregulated DEGs were enriched in eye development functions at 35 dpf. The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed enrichment of downregulated DEGs in focal adhesion and extracellular matrix (ECM)-receptor interaction pathways at both stages. Neuronal development DEGs were mainly downregulated at both stages, while synaptic signaling DEGs were upregulated at 35 dpf. Crossing cdkl5-/- mutants with the Hb9:GFP transgenic line showed fewer motor neuron cells with shorter axons compared to the wild type, which may explain the impaired motor phenotype observed in zebrafish and CDD patients. Moreover, we identified key downregulated DEGs related to cartilage development at both stages and bone development at 35 dpf, potentially explaining the skeletal defects seen in zebrafish and CDD individuals. In conclusion, Cdkl5 loss in zebrafish leads to dysregulation of genes involved in CDKL5-associated functions in mammals, providing new insights into its less studied functions and phenotypes.
Keywords: CDKL5 deficiency disorder; Cdkl5; RNA-seq; transcriptomic; zebrafish model.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Rusconi L., Salvatoni L., Giudici L., Bertani I., Kilstrup-Nielsen C., Broccoli V., Landsberger N. CDKL5 expression is modulated during neuronal development and its subcellular distribution is tightly regulated by the C-terminal tail. J. Biol. Chem. 2008;283:30101–30111. doi: 10.1074/jbc.M804613200. - DOI - PMC - PubMed
-
- Ricciardi S., Ungaro F., Hambrock M., Rademacher N., Stefanelli G., Brambilla D., Sessa A., Magagnotti C., Bachi A., Giarda E., et al. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat. Cell Biol. 2012;14:911–923. doi: 10.1038/ncb2566. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

