Post-Transcriptional Regulation of the MiaA Prenyl Transferase by CsrA and the Small RNA CsrB in Escherichia coli
- PMID: 40649849
- PMCID: PMC12249760
- DOI: 10.3390/ijms26136068
Post-Transcriptional Regulation of the MiaA Prenyl Transferase by CsrA and the Small RNA CsrB in Escherichia coli
Abstract
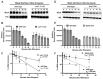
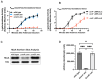
MiaA is responsible for the addition of the isopentyl modification to adenine 37 in the anticodon stem loop of specific tRNAs in Escherichia coli. Mutants in miaA have pleotropic effects on the cell in E. coli and play a role in virulence gene regulation. In addition, MiaA is necessary for stress response gene expression by promoting efficient decoding of UUX-leucine codons, and genes with elevated UUX-leucine codons may be a regulatory target for i6A-modified tRNAs. Understanding the temporal nature of the i6A modification status of tRNAs would help us determine the regulatory potential of MiaA and its potential interplay with leucine codon frequency. In this work, we set out to uncover additional information about the synthesis of the MiaA. MiaA synthesis is primarily driven at the transcriptional level from multiple promoters in a complex operon. However, very little is known about the post-transcriptional regulation of MiaA, including the role of sRNAs in its synthesis. To determine the role of small RNAs (sRNAs) in the regulation of miaA, we constructed a chromosomal miaA-lacZ translational fusion driven by the arabinose-responsive PBAD promoter and used it to screen against an Escherichia coli sRNA library (containing sRNAs driven by the IPTG-inducible PLac promoter). Our genetic screen and quantitative β-galactosidase assays identified CsrB and its cognate protein CsrA as potential regulators of miaA expression in E. coli. Consistent with our hypothesis that CsrA regulates miaA post-transcriptional gene expression through binding to the miaA mRNA 5' UTR, and CsrB binds and regulates miaA post-transcriptional gene expression through sequestration of CsrA levels, a deletion of csrA significantly reduced expression of the reporter fusion as well as reducing miaA mRNA levels. These results suggest that under conditions where CsrA is inhibited, miaA mRNA translation and thus MiaA-dependent tRNA modification may be limited.
Keywords: RNA binding protein; RNA processing; small RNA; tRNA modification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
CsrA-mediated regulation of a virulence switch in Acinetobacter baumannii.mBio. 2025 Apr 9;16(4):e0405824. doi: 10.1128/mbio.04058-24. Epub 2025 Feb 25. mBio. 2025. PMID: 39998216 Free PMC article.
-
TrmL and TusA Are Necessary for rpoS and MiaA Is Required for hfq Expression in Escherichia coli.Biomolecules. 2017 May 4;7(2):39. doi: 10.3390/biom7020039. Biomolecules. 2017. PMID: 28471404 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Hfq and sRNA 179 Inhibit Expression of the Pseudomonas aeruginosa cAMP-Vfr and Type III Secretion Regulons.mBio. 2020 Jun 16;11(3):e00363-20. doi: 10.1128/mBio.00363-20. mBio. 2020. PMID: 32546612 Free PMC article.
-
123I-MIBG scintigraphy and 18F-FDG-PET imaging for diagnosing neuroblastoma.Cochrane Database Syst Rev. 2015 Sep 29;2015(9):CD009263. doi: 10.1002/14651858.CD009263.pub2. Cochrane Database Syst Rev. 2015. PMID: 26417712 Free PMC article.
References
-
- Connolly D.M., Winkler M.E. Genetic and physiological relationships among the miaA gene, 2-methylthio-N6-(delta 2-isopentenyl)-adenosine tRNA modification, and spontaneous mutagenesis in Escherichia coli K-12. J. Bacteriol. 1989;171:3233–3246. doi: 10.1128/jb.171.6.3233-3246.1989. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

