T-Cadherin (CDH13) and Non-Coding RNAs: The Crosstalk Between Health and Disease
- PMID: 40649905
- PMCID: PMC12250294
- DOI: 10.3390/ijms26136127
T-Cadherin (CDH13) and Non-Coding RNAs: The Crosstalk Between Health and Disease
Abstract
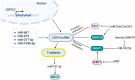
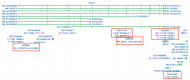
T-cadherin (CDH13) is an atypical, glycosyl-phosphatidylinositol-anchored cadherin with functions ranging from axon guidance and vascular patterning to adipokine signaling and cell-fate specification. Originally identified as a homophilic cue for migrating neural crest cells, projecting axons, and growing blood vessels, it later emerged as a dual metabolic receptor for cardioprotective high-molecular-weight adiponectin and atherogenic low-density lipoproteins. We recently showed that mesenchymal stem/stromal cells lacking T-cadherin are predisposed to adipogenesis, underscoring its role in lineage choice. Emerging evidence indicates that CDH13 expression and function are fine-tuned by non-coding RNAs (ncRNAs). MiR-199b-5p, miR-377-3p, miR-23a/27a/24-2, and the miR-142 family directly bind CDH13 3'-UTR or its epigenetic regulators, affecting transcription or accelerating decay. Long non-coding RNAs (lncRNAs), including antisense transcripts CDH13-AS1/AS2, brain-restricted FEDORA, and context-dependent LINC00707 and UPAT, either sponge these miRNAs or recruit DNMT/TET enzymes to the CDH13 promoter. Circular RNAs (circRNAs), i.e.circCDH13 and circ_0000119, can add a third level of complexity by sequestering miRNA repressors or boosting DNMT1. Collectively, this ncRNA circuitry regulates T-cadherin across cardiovascular, metabolic, oncogenic, and neurodegenerative conditions. This review integrates both experimentally validated data and in silico predictions to map the ncRNA-CDH13 crosstalk between health and disease, opening new avenues for biomarker discovery and RNA-based therapeutics.
Keywords: CDH13; T-cadherin; miRNA; non-coding RNA; regulation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

