Molecular Study from the Signaling Pathways of Four Potential asthma triggers: AKT1, MAPK13, STAT1, and TLR4
- PMID: 40650018
- PMCID: PMC12249927
- DOI: 10.3390/ijms26136240
Molecular Study from the Signaling Pathways of Four Potential asthma triggers: AKT1, MAPK13, STAT1, and TLR4
Abstract
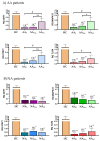
Asthma is a chronic and heterogeneous inflammatory airway disease with diverse clinical endotypes and limited curative treatment options. Recent systems biology analyses identified four potential molecular triggers-AKT1, MAPK13, STAT1, and TLR4-as candidate regulators of asthma-associated signaling pathways. This study aimed to validate the expression of these four proteins and their downstream signaling elements in peripheral blood mononuclear cells (PBMCs) from patients with allergic asthma (AA), nonallergic asthma (NA), and healthy controls (HC), to explore their potential as biomarkers or therapeutic targets. For that, PBMC samples were collected from 45 AA patients, 17 NA patients, and 15 HC subjects. Gene and protein expression of AKT1, MAPK13, STAT1, and TLR4 were quantified using RT-qPCR and Western blotting. Expression patterns were compared across groups and stratified by asthma severity. Correlations with clinical parameters (FEV1, FVC, FeNO, IgE, eosinophil counts) and treatment regimens were also assessed. All four target genes showed significantly reduced expression in asthma patients compared to controls (p < 0.001), with the most marked downregulation in NA patients. At the protein level, MAPK13 and TLR4 showed significant differential expression. Stratification by severity revealed a stepwise reduction in gene expression in AA patients, correlating with disease severity, whereas NA patients showed uniformly low expression regardless of severity. Multiple pathway-related genes, including RELA, SMAD3, NFATC1, and ALOX5, were also downregulated, particularly in NA patients. Notably, differential correlations were observed between gene expression and lung function parameters in AA vs. NA groups. In conclusion, this study supports the potential involvement of AKT1, MAPK13, STAT1, and TLR4 in asthma pathogenesis and highlights differences between allergic and nonallergic asthma at the molecular level. These proteins and their associated pathways may serve as future targets for biomarker development or endotype-specific therapies. Further studies in larger and more diverse cohorts, including functional validation, are warranted.
Keywords: AKT1; MAPK13; STAT1; TLR4; allergic asthma (AA); asthma; biomarkers; gene expression; nonallergic asthma (NA); peripheral blood mononuclear cells (PBMCs); signaling pathways.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- GINA. Global Initiative for Asthma Global Strategy for Asthma Management and Prevention. 2021. [(accessed on 28 May 2024)]. Available online: http://www.ginasthma.org/
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

