Salivary Metabolomics Discloses Metabolite Signatures of Oral Leukoplakia with and Without Dysplasia
- PMID: 40650295
- PMCID: PMC12250079
- DOI: 10.3390/ijms26136519
Salivary Metabolomics Discloses Metabolite Signatures of Oral Leukoplakia with and Without Dysplasia
Abstract
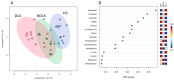
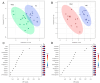
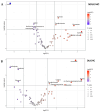
Leukoplakia is a condition marked by white patches on the inner surfaces of the oral cavity. Its potential to progress to oral squamous cell carcinoma underscores the need for effective screening and early diagnosis procedures. We employed NMR-based salivary and tissue metabolomics to identify potential biomarkers for leukoplakia and dysplastic leukoplakia. Univariate and multivariate methods were used to evaluate the NMR-derived metabolite concentrations. The salivary metabolite profile of leukoplakia exhibited specific alterations compared to healthy controls. These metabolic changes were more pronounced in cases of dysplastic lesions. Multivariate ROC curve analysis, based on a selection of salivary metabolites, ascribed high diagnostic accuracy to the models that discriminate between dysplastic and healthy cases. However, NMR analysis of tissue biopsies was ineffective in extracting metabolic signatures to differentiate between lesional, peri-lesional, and healthy tissues. Our pilot study employing a metabolomics-based approach led to the development of salivary models that represent a complementary strategy for clinically detecting leukoplakia. However, larger-scale validation is required to fully evaluate their diagnostic potential and to effectively stratify leukoplakia patients according to dysplasia status.
Keywords: dysplasia; leukoplakia; malignant transformation; salivary diagnostics; salivary metabolomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Metabolomics-based predictive biomarkers of oral cancer and its severity in human patients from North India using saliva.Mol Omics. 2025 Jul 7;21(4):270-281. doi: 10.1039/d4mo00166d. Mol Omics. 2025. PMID: 39945460
-
A Deep Learning System to Predict Epithelial Dysplasia in Oral Leukoplakia.J Dent Res. 2024 Nov;103(12):1218-1226. doi: 10.1177/00220345241272048. Epub 2024 Oct 9. J Dent Res. 2024. PMID: 39382109
-
Interventions for treating oral leukoplakia to prevent oral cancer.Cochrane Database Syst Rev. 2016 Jul 29;7(7):CD001829. doi: 10.1002/14651858.CD001829.pub4. Cochrane Database Syst Rev. 2016. PMID: 27471845 Free PMC article.
-
Plasma metabolites associated with endometriosis in adolescents and young adults.Hum Reprod. 2025 May 1;40(5):843-854. doi: 10.1093/humrep/deaf040. Hum Reprod. 2025. PMID: 40107296
-
Diagnostic tests for oral cancer and potentially malignant disorders in patients presenting with clinically evident lesions.Cochrane Database Syst Rev. 2015 May 29;2015(5):CD010276. doi: 10.1002/14651858.CD010276.pub2. Cochrane Database Syst Rev. 2015. Update in: Cochrane Database Syst Rev. 2021 Jul 20;7:CD010276. doi: 10.1002/14651858.CD010276.pub3. PMID: 26021841 Free PMC article. Updated.
References
-
- Cui Y., Yang M., Zhu J., Zhang H., Duan Z., Wang S., Liao Z., Liu W. Developments in diagnostic applications of saliva in human organ diseases. Med. Nov. Technol. Devices. 2022;13:100115. doi: 10.1016/j.medntd.2022.100115. - DOI
-
- Khurshid Z., Warsi I., Moin S.F., Slowey P.D., Latif M., Zohaib S., Zafar M.S. Biochemical analysis of oral fluids for disease detection. Adv. Clin. Chem. 2021;100:205–253. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

