An mRNA Vaccine Targeting the C-Terminal Region of P1 Protein Induces an Immune Response and Protects Against Mycoplasma pneumoniae
- PMID: 40650312
- PMCID: PMC12249837
- DOI: 10.3390/ijms26136536
An mRNA Vaccine Targeting the C-Terminal Region of P1 Protein Induces an Immune Response and Protects Against Mycoplasma pneumoniae
Abstract
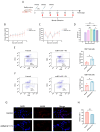
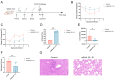
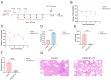
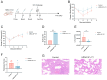
Mycoplasma pneumoniae, a cell wall-deficient pathogen, primarily affects children and adolescents, causing Mycoplasma pneumoniae pneumonia (MPP). Following the relaxation of non-pharmaceutical interventions (NPIs) post COVID-19, there has been a global increase in MPP cases and macrolide-resistant strains. Vaccination against M. pneumoniae is being explored as a promising approach to reduce infections, limit antibiotic misuse, and prevent the emergence of drug-resistant variants. We developed an mRNA vaccine, mRNA-SP+P1, incorporating a eukaryotic signal peptide (tissue-type plasminogen activator signal peptide) fused to the C-terminal region of the P1 protein. Targeting amino acids 1288 to 1518 of the P1 protein, the vaccine was administered intramuscularly to BALB/c mice in a three-dose regimen. To evaluate immunogenicity, we quantified anti-P1 IgG antibody titers using enzyme-linked immunosorbent assays (ELISAs) and assessed cellular immune responses by analyzing effector memory T cell populations using flow cytometry. We also tested the functional activity of vaccine-induced sera for their ability to inhibit adhesion of the ATCC M129 strain to KMB17 cells. The vaccine's protective efficacy was assessed against the ATCC M129 strain and its cross-protection against the ST3-resistant strain. Transcriptomic analysis was conducted to investigate gene expression changes in peripheral blood, aiming to uncover mechanisms of immune modulation. The mRNA-SP+P1 vaccine induces P1 protein-specific IgG antibodies and an effector memory T-cell response in BALB/c mice. Adhesion inhibition assays demonstrated that serum from vaccinated mice attenuatesthe adhesion ability of ATCC M129 to KMB17 cells. Furthermore, three doses of the vaccine confer significant and long-lasting, though partial, protection against the ATCC M129 strain and partial cross-protection against the ST3 drug-resistant strain. Transcriptome analysis revealed significant gene expression changes in peripheral blood, confirming the vaccine's capacity to elicit an immune response from the molecular level. Our results indicate that the mRNA-SP+P1 vaccine appears to be an effective vaccine candidate against the prevalence of Mycoplasma pneumoniae.
Keywords: ATCC M129 strain; Mycoplasma pneumoniae; P1 protein; ST3 drug-resistant strain; mRNA vaccine.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Protective immunity induced by a novel P1 adhesin C-terminal anchored mRNA vaccine against Mycoplasma pneumoniae infection in BALB/c mice.Microbiol Spectr. 2025 Mar 4;13(3):e0214024. doi: 10.1128/spectrum.02140-24. Epub 2025 Jan 20. Microbiol Spectr. 2025. PMID: 39831768 Free PMC article.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
Evolution and characteristics of tetracycline resistance in Mycoplasma pneumoniae.Microbiol Spectr. 2025 Jul;13(7):e0339824. doi: 10.1128/spectrum.03398-24. Epub 2025 May 23. Microbiol Spectr. 2025. PMID: 40407312 Free PMC article.
-
Antibiotics for community-acquired lower respiratory tract infections secondary to Mycoplasma pneumoniae in children.Cochrane Database Syst Rev. 2012 Sep 12;(9):CD004875. doi: 10.1002/14651858.CD004875.pub4. Cochrane Database Syst Rev. 2012. Update in: Cochrane Database Syst Rev. 2015 Jan 08;1:CD004875. doi: 10.1002/14651858.CD004875.pub5. PMID: 22972079 Updated.
-
Antibiotics for community-acquired lower respiratory tract infections secondary to Mycoplasma pneumoniae in children.Cochrane Database Syst Rev. 2010 Jul 7;(7):CD004875. doi: 10.1002/14651858.CD004875.pub3. Cochrane Database Syst Rev. 2010. Update in: Cochrane Database Syst Rev. 2012 Sep 12;(9):CD004875. doi: 10.1002/14651858.CD004875.pub4. PMID: 20614439 Updated.
References
-
- Lee E., Kim C.H., Lee Y.J., Kim H.B., Kim B.S., Kim H.Y., Kim Y., Kim S., Park C., Seo J.H., et al. Annual and seasonal patterns in etiologies of pediatric community-acquired pneumonia due to respiratory viruses and Mycoplasma pneumoniae requiring hospitalization in South Korea. BMC Infect. Dis. 2020;20:132. doi: 10.1186/s12879-020-4810-9. - DOI - PMC - PubMed
-
- Zhang Y., Su C., Zhang Y., Ding S., Yan X., Zhang J., Tao Z. Epidemiological and clinical characteristics of hospitalized pediatric patients with Mycoplasma pneumoniae pneumonia before and after the COVID-19 pandemic in China: A retrospective multicenter study. BMC Infect. Dis. 2025;25:18. doi: 10.1186/s12879-024-10370-8. - DOI - PMC - PubMed
-
- Wang R.S., Wang S.Y., Hsieh K.S., Chiou Y.H., Huang I.F., Cheng M.F., Chiou C.C. Necrotizing pneumonitis caused by Mycoplasma pneumoniae in pediatric patients: Report of five cases and review of literature. Pediatr. Infect. Dis. J. 2004;23:564–567. doi: 10.1097/01.inf.0000130074.56368.4b. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

