Single-cell RNA sequencing and multi-omics analysis of prognosis-related staging in papillary thyroid cancer
- PMID: 40650680
- PMCID: PMC12255609
- DOI: 10.1007/s00262-025-04101-4
Single-cell RNA sequencing and multi-omics analysis of prognosis-related staging in papillary thyroid cancer
Abstract
Background: Papillary thyroid cancer (PTC) is the most common thyroid cancer, but current molecular features inadequately stratify its risk. Whether distinct underlying mechanisms can further classify PTC and improve prognostic precision remains unclear.
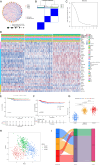
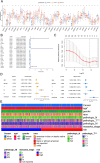
Methods: We integrated single-cell RNA sequencing data (158,577 cells from 11 PTC patients; GEO: GSE184362) with bulk-RNA sequencing data from The Cancer Genome Atlas Thyroid Carcinoma (TCGA-THCA) cohort (501 patients). Multi-omics analyses were employed to elucidate PTC heterogeneity, identify malignant cell differentiation and prognosis-related genes (MCD&PRGs), and construct a novel molecular classification, the Oncogenic Signature Of Papillary Thyroid Carcinoma Classification (OSPTCC). A prognostic risk score was developed, and the classification's prognostic relevance was further explored in an independent institutional cohort using qRT-PCR.
Results: Single-cell analysis revealed three malignant cell differentiation states (PTC1-3) and a 34-gene signature (MCD&PRGs). This formed the basis of our Oncogenic Signature Of Papillary Thyroid Carcinoma Classification (OSPTCC), defining three subtypes: Inflammation-associated (IPTCC), BRAF/autophagy-related (BAPTCC), and lipid metabolism-related (LPTCC). These subtypes showed distinct molecular profiles and significantly different progression-free survival (IPTCC poorest, P = 0.044). A 7-gene risk score derived from MCD&PRGs independently predicted prognosis (multivariate HR = 21.511, P < 0.001). qRT-PCR validation in an independent cohort (n = 48) using key markers (DEPTOR, APOE, APOC1) confirmed that OSPTCC-based risk stratification correlated with adverse clinical features, including higher recurrence rates in the high-risk group (P = 0.007).
Conclusions: This study introduces OSPTCC, a prognostically significant molecular classification for PTC based on tumor cell differentiation states. The identified subtypes, characterized by distinct biological mechanisms, provide deeper insights into PTC's molecular pathology and offer a framework for improved risk stratification and potential precision therapies.
Keywords: Molecular subtypes; Multi-omics; Papillary thyroid cancer; Single-cell RNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: N/A Informed Consent: N/A Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Leenhardt L, Grosclaude P, Chérié-Challine L (2004) Increased incidence of thyroid carcinoma in france: a true epidemic or thyroid nodule management effects? Report from the French Thyroid Cancer Committee. Thyroid 14(12):1056–1060 - PubMed
-
- Wiltshire JJ et al (2016) Systematic Review of Trends in the Incidence Rates of Thyroid Cancer. Thyroid 26(11):1541–1552 - PubMed
-
- Davies L, Welch HG (2014) Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg 140(4):317–322 - PubMed
-
- Ahn HS, Kim HJ, Welch HG (2014) Korea’s thyroid-cancer “epidemic”–screening and overdiagnosis. N Engl J Med 371(19):1765–1767 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

