Identification of a novel complex variant in a patient involving the α-globin gene cluster by third-generation sequencing
- PMID: 40650714
- PMCID: PMC12334446
- DOI: 10.1007/s00277-025-06488-7
Identification of a novel complex variant in a patient involving the α-globin gene cluster by third-generation sequencing
Abstract
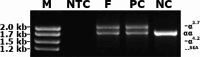
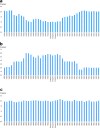
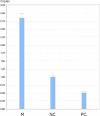
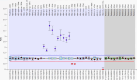
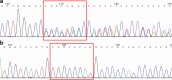
Various methods are available to detect common deletions and mutations of genes related to thalassemia, including gap-polymerase chain reaction (Gap-PCR), next-generation sequencing (NGS), multiplex ligation-dependent probe amplification (MLPA), and quantitative real-time polymerase chain reaction (qRT-PCR). Unequal crossover during the recombination of α1 and α2 hemoglobin can be detected but hardly accurately defined by above-mentioned technologies. A couple with abnormal hematological test results arrived at our department for genetic consultation. Preliminary analysis using NGS revealed a 3.7 kb (chr16:223462-227311) heterozygote deletion in the wife and nearly six copies in the chr16:223462-227311 (GRCh37/hg19) region of the husband. Further Gap-PCR results for the wife were consistent with the NGS results. MLPA and qRT-PCR were performed to detect the potential extra copies of the α-globin gene of the husband. The analyses simultaneously showed that the copy numbers of HBA1/HBA2 genes were nearly six. A specialized primer of the α-globin gene was designed to elucidate the structure of the 4.2 or 3.7 kb repeats of the husband. Third-generation sequencing (TGS) revealed the existence of the four extra tandem duplications of 3.7 kb in DNA strand 1 (chr16:173302-177106, hg38) of the α-globin gene and the existence of a heterozygous c.301-31_301-24delinsG insertion and deletion (InDel) in DNA strand 2 in the HBA1 gene (chr16:177252-177259, hg38). These results were confirmed by Sanger sequencing. Integrative Genomics Viewer analysis of the BAM files of NGS detected a low-level (reference/alternative, 0.19%) heterozygous InDel (c.301-31_301-24delinsG) in HBA1. Compared to traditional methods, TGS was better able to detect variants accurately and find rare genotypes and rearrangements of the α-globin gene cluster.
Keywords: Multiple copies of α-globin gene cluster; Thalassemia; Third-generation sequencing; Traditional methods.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics statement: The studies involving humans were approved by the Medical Ethics Committee of the West China Second University Hospital of Sichuan University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individuals for the publication of any potentially identifiable images or data included in this article. Informed consent: The patient and her family signed informed consent. Competing interests: The authors declare no competing interests.
Figures

References
-
- Kattamis A, Kwiatkowski JL, Aydinok Y (2022) Thalassaemia. Lancet 399:2310–2324. 10.1016/S0140-6736(22)00536-0 - PubMed
-
- Long J, Liu E (2021) The carriage rates of αααanti3.7, αααanti4.2, and HKαα in the population of guangxi, China measured using a rapid detection qPCR system to determine CNV in the α-globin gene cluster. Gene 768:145296. 10.1016/j.gene.2020.145296 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

