CRISPR screening approaches in breast cancer research
- PMID: 40650785
- PMCID: PMC12255600
- DOI: 10.1007/s10555-025-10275-1
CRISPR screening approaches in breast cancer research
Abstract
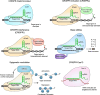
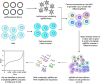
The emergence of CRISPR-Cas9 technology has transformed functional genomics, offering unmatched opportunities to dissect and understand biological pathways and identify novel therapeutic targets in cancer. Breast cancer is a complex, heterogeneous disease and remains a major cause of morbidity and mortality in women, particularly when diagnosed at advanced or metastatic stages where effective treatments are limited. High-throughput CRISPR screening is undoubtedly a powerful tool to discover novel drug targets, uncover synthetic lethal interactions, and identify vulnerabilities in cancer. This review focuses on advances in our understanding of breast cancer developed through CRISPR-based screening technology, particularly in identifying drivers of breast cancer progression, growth, and metastasis, as well as in identifying potential new therapeutic targets and combination therapies. We discuss recent discoveries, current challenges, and limitations of this approach and explore how advancements in CRISPR technology could have a profound impact on the future of breast cancer treatment.
Keywords: Breast cancer; CRISPR; Drug discovery; Drug resistance; Functional genomics; Novel therapies; Screening.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: Not applicable. Informed consent: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Genome-scale CRISPR-Cas9 screening in stem cells: theories, applications and challenges.Stem Cell Res Ther. 2024 Jul 19;15(1):218. doi: 10.1186/s13287-024-03831-z. Stem Cell Res Ther. 2024. PMID: 39026343 Free PMC article. Review.
-
Exploring synthetic lethality in cancer therapy: CRISPR-Cas9 technology offers new hope.Biochim Biophys Acta Rev Cancer. 2025 Sep;1880(4):189370. doi: 10.1016/j.bbcan.2025.189370. Epub 2025 Jun 13. Biochim Biophys Acta Rev Cancer. 2025. PMID: 40516634 Review.
-
Genome-wide CRISPR Screening Identifies NFκB and c-MET as Druggable Targets to Sensitize Lenvatinib Treatment in Hepatocellular Carcinoma.Cell Mol Gastroenterol Hepatol. 2025;19(7):101502. doi: 10.1016/j.jcmgh.2025.101502. Epub 2025 Mar 20. Cell Mol Gastroenterol Hepatol. 2025. PMID: 40120675 Free PMC article.
-
CRISPR/dCas-mediated counter-silencing: reprogramming dCas proteins into antagonists of xenogeneic silencers.mBio. 2025 Jul 9;16(7):e0038225. doi: 10.1128/mbio.00382-25. Epub 2025 May 28. mBio. 2025. PMID: 40434115 Free PMC article.
-
Interventions for promoting habitual exercise in people living with and beyond cancer.Cochrane Database Syst Rev. 2018 Sep 19;9(9):CD010192. doi: 10.1002/14651858.CD010192.pub3. Cochrane Database Syst Rev. 2018. PMID: 30229557 Free PMC article.
References
-
- Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., & Jemal, A. (2024). Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians,74(3), 229–263. - PubMed
-
- Harbeck, N., Penault-Llorca, F., Cortes, J., Gnant, M., Houssami, N., Poortmans, P., et al. (2019). Breast cancer. Nature Reviews Disease Primers,5(1), 66. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

