BLIMP1 controls GC B cell expansion and exit through regulating cell cycle progression and key transcription factors BCL6 and IRF4
- PMID: 40650910
- PMCID: PMC12356087
- DOI: 10.1016/j.celrep.2025.115977
BLIMP1 controls GC B cell expansion and exit through regulating cell cycle progression and key transcription factors BCL6 and IRF4
Abstract
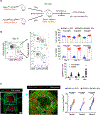
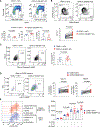
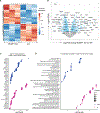
In B cells, BLIMP1 is required for plasma cell differentiation. BLIMP1 is also expressed in some germinal center (GC) B cells (GCBC), yet the role of BLIMP1 in GCBC is not understood. Here we generated mixed bone marrow (BM) chimeric mice using Prdm1+/+ CD19Cre and Prdm1fl/fl CD19Cre BM, allowing us to examine the cell-intrinsic functions of BLIMP1 in GCBC, independent of antibody or antigen levels. Strikingly, BLIMP1-deficient B cells quickly dominate GCs and persist for a much longer time compared with wild-type cells. BLIMP1 deficiency promotes positive selection of GCBCs and enhances cell-cycle progression. Additionally, BLIMP1 deficiency alters class switching and memory B cell generation from GCBCs. Mechanistically, BLIMP1-deficient GCBCs fail to downregulate BCL6 and to upregulate IRF4, indicating that BLIMP1 controls the expression of these transcription factors that mediate exit from the GC. These studies revealed unique functions of BLIMP1 in regulating GCBC responses that impact long-lived immune compartments.
Keywords: B cell; BCL6; Blimp1; CP: Immunology; germinal center; memory B cells; plasma cells; transcription factors.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.H.K. receives consulting fees from Peraton.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

