A-to-I mRNA editing recodes hundreds of genes in dozens of species and produces endogenous protein isoforms in bacteria
- PMID: 40650968
- PMCID: PMC12255301
- DOI: 10.1093/nar/gkaf656
A-to-I mRNA editing recodes hundreds of genes in dozens of species and produces endogenous protein isoforms in bacteria
Abstract
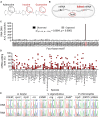
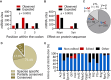
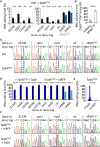
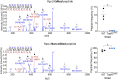
Adenosine-to-inosine (A-to-I) messenger RNA (mRNA) editing can affect the sequence and function of translated proteins and has been extensively investigated in eukaryotes. However, the prevalence of A-to-I mRNA editing in bacteria, its governing regulatory principles, and its biological significance are poorly understood. Here, we show that A-to-I mRNA editing occurs in hundreds of transcripts across dozens of gammaproteobacterial species, with most edits predicted to recode protein sequences. Furthermore, we reveal conserved regulatory determinants controlling editing across gammaproteobacterial species. Using Acinetobacter baylyi as a model, we show that mutating TadA, the mediating enzyme, reduces editing across all sites. Conversely, overexpressing TadA resulted in the editing of >300 transcripts, attesting to the editing potential of TadA. Notably, we show for the first time, at the protein level, that normal levels of A-to-I mRNA editing lead to wild-type bacteria expressing two protein isoforms from a single gene. Finally, we show that a TadA mutant with deficient editing activity does not grow at high temperatures, suggesting that RNA editing has a functional role in bacteria. Our work reveals that A-to-I mRNA editing in bacteria is widespread and has the potential to reshape the bacterial transcriptome and proteome.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

