Design of mismatch closure for enhanced specificity in DNA strand displacement reactions
- PMID: 40650970
- PMCID: PMC12255293
- DOI: 10.1093/nar/gkaf660
Design of mismatch closure for enhanced specificity in DNA strand displacement reactions
Abstract
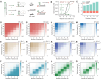
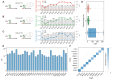
Specific and sensitive DNA hybridization plays a key role in biotechnology, nanotechnology, and medical technology. However, traditional DNA hybridization-based strategies often require careful tuning of the binding affinity of the probe to attain a trade-off between specificity and sensitivity. Herein, we proposed energy barrier-gated dynamic selectivity to overcome this limitation. The mismatch closure-mediated strand displacement reaction (mcSDR) induces structural constraints through helper strand binding at mismatched sites, resulting in the displacement of the mismatch target requires overcoming an additional activation energy barrier, whereas the perfect match target proceeds via a normal pathway. The mcSDR has been thermodynamically and kinetically demonstrated to be able to balance specificity and sensitivity simultaneously. The energy barrier height can be programmably adjusted by design of helper strand and works in synergy with the toehold exchange strategy to achieve multi-parameter optimization. The superior properties of the mcSDR facilitated the identification of 12 mutation types exhibits excellent specificity in 28 clinically relevant single nucleotide variations. By combining polymerase chain reaction, mutations with an abundance of 0.1% were successfully detected in plasmid samples, and a triple mcSDR was successfully constructed. Clinical validation of 95 glioma and 93 colorectal cancer samples showed that IDH1 and KRAS mutations were 100% consistent with Sanger sequencing. The energy barrier-driven identification mechanism and operational simplicity of mcSDR make it promising for wide applications in biomedical research, molecular diagnosis, and precision medicine.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures





References
-
- Seeman NC, Sleiman HF DNA nanotechnology. Nat Rev Mater. 2017; 3:17068. 10.1038/natrevmats.2017.68. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

