Dynamic gene regulatory network inference from single-cell data using optimal transport
- PMID: 40650986
- PMCID: PMC12352743
- DOI: 10.1093/bioinformatics/btaf394
Dynamic gene regulatory network inference from single-cell data using optimal transport
Abstract
Motivation: Modelling gene expression is a central problem in systems biology. Single-cell technologies have revolutionized the field by enabling sequencing at the resolution of individual cells. This results in a much richer data compared to what is obtained by bulk technologies, offering new possibilities and challenges for gene regulatory network inference.
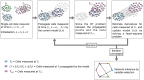
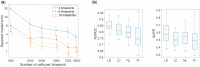
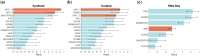
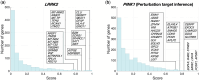
Results: In this work, we introduce GRIT (gene regulation inference by transport)-a method to fit a differential equation model and to infer gene regulatory networks from single-cell data using the theory of optimal transport. The idea consists in tracking the evolution of the cell distribution over time and finding the system whose temporal marginals minimize the transport cost with the observations. GRIT is finally used to identify genes and pathways affected by two Parkinson's disease associated mutations.
Availability and implementation: Matlab implementation of the method and code for data generation are at gitlab.com/uniluxembourg/lcsb/systems-control/grit together with a user guide. A snapshot of the code used for the results of this article is at doi: 10.5281/zenodo.15582432.
© The Author(s) 2025. Published by Oxford University Press.
Figures
References
-
- Aalto A, Gonçalves J. Linear system identification from ensemble snapshot observations. In: 2019 IEEE 58th Conference on Decision and Control (CDC), Nice, France. IEEE Control Systems Society, 2019, 7554–9. 10.1109/CDC40024.2019.9029334 - DOI
-
- Aalto A, Lamoline F, Gonçalves J. Linear system identifiability from single-cell data. Syst Control Lett 2022;165:105287. 10.1016/j.sysconle.2022.105287 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

