Interpreting ribosome dynamics during mRNA translation
- PMID: 40651611
- PMCID: PMC12340396
- DOI: 10.1016/j.jbc.2025.110469
Interpreting ribosome dynamics during mRNA translation
Abstract
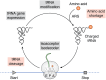
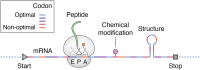
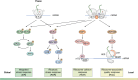
Translation takes a central position in gene expression, and its swift response to environmental stress is evolutionarily conserved. Upon chemical damage to the messenger RNA (mRNA) or the lack of building blocks, the ribosome stalls during elongation and halts the production line. Even under normal growth conditions, the translation machinery encounters constant hindrances such as varied codon composition or nascent chains with distinct features. However, it is challenging to define these kinetics experimentally, partly due to the inherent variations of ribosome behavior during mRNA translation. To ensure the flow of ribosomal traffic, cells employ several mechanisms to circumvent the traffic jam. When the roadblock is not resolved timely, trailing ribosomes can collide with stalled ribosomes. However, the boundary between physiological queuing and pathological collision is often blurred, representing a fundamental gap in our understanding of ribosome dynamics. To cope with translational barriers, several signaling pathways are activated to adjust the rate of global translation and rescue the local stalled ribosome. Deficiencies in cellular response to translational stress have been associated with a wide array of human diseases. In this review, we focus on fundamental aspects of the ribosome dynamics during mRNA translation. We provide an overview of causes, outcomes, and cellular responses to ribosome stalling and collision on mRNA. We highlight questions that may clarify the biological roles of distinct ribosome behavior during mRNA translation and emphasize the mechanistic connection between altered ribosome dynamics and human diseases.
Keywords: codon; mRNA; protein synthesis; ribosome; stress response; tRNA; translation.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Quantification of elongation stalls and impact on gene expression in yeast.RNA. 2023 Dec;29(12):1928-1938. doi: 10.1261/rna.079663.123. Epub 2023 Oct 2. RNA. 2023. PMID: 37783489 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Translation Rates and Protein Folding.J Mol Biol. 2024 Jul 15;436(14):168384. doi: 10.1016/j.jmb.2023.168384. Epub 2023 Dec 6. J Mol Biol. 2024. PMID: 38065274 Free PMC article. Review.
-
What, where, and how: Regulation of translation and the translational landscape in plants.Plant Cell. 2024 May 1;36(5):1540-1564. doi: 10.1093/plcell/koad197. Plant Cell. 2024. PMID: 37437121 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

