Genome mining for the discovery of peptide halogenases and their biochemical characterization
- PMID: 40651833
- PMCID: PMC12255868
- DOI: 10.1016/bs.mie.2025.02.006
Genome mining for the discovery of peptide halogenases and their biochemical characterization
Abstract
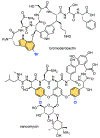
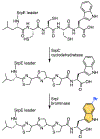
While halogenation is one of the most versatile C-H functionalization strategy, regiospecific halogenation of peptides and proteins is outside the purview of traditional chemical catalysis. Enzymes that participate in the biosynthesis of ribosomally synthesized and post-translationally modified peptides and proteins can bridge this gap and offer a biocatalytic route for residue-specific incorporation of halogen handles onto amino acid side chains. Protocols described herein provide a guided approach for the discovery of peptide halogenases in the context of natural product biosynthetic gene clusters, and the preliminary reconstitution of their activity using a bacterial heterologous host. Also described are mass spectrometry-based analytical procedures and data analysis workflows that allow for deconvolution of halide specificity and preliminary insights into peptidic natural product biosynthetic schemes. As the available genomic data expands at a rapid rate, the methodology described here will enable the discovery and characterization of new halogenases that can be valuable partners in chemoenzymatic diversification of peptides and proteins.
Keywords: Genome mining; Halogenase; Mass spectrometry; RiPPs.
Copyright © 2025. Published by Elsevier Inc.
Figures


Similar articles
-
Characterization of an Iterative Halogenase Acting on Ribosomal Peptides Underlies the Combinatorial Biosynthesis Logic of Lasso Peptides.J Nat Prod. 2025 Mar 28;88(3):650-661. doi: 10.1021/acs.jnatprod.4c01199. Epub 2025 Feb 20. J Nat Prod. 2025. PMID: 39976742
-
Exploring ribosomally synthesized and post-translationally modified peptides through SPECO-based genome mining.Methods Enzymol. 2025;717:67-87. doi: 10.1016/bs.mie.2025.04.005. Epub 2025 May 24. Methods Enzymol. 2025. PMID: 40651835
-
Genome mining and in vivo characterization of triceptides.Methods Enzymol. 2025;717:119-140. doi: 10.1016/bs.mie.2025.04.002. Epub 2025 May 28. Methods Enzymol. 2025. PMID: 40651822
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Plant peptides - redefining an area of ribosomally synthesized and post-translationally modified peptides.Nat Prod Rep. 2024 Jul 17;41(7):1020-1059. doi: 10.1039/d3np00042g. Nat Prod Rep. 2024. PMID: 38411572 Free PMC article. Review.
References
-
- Böhringer N, Kramer J, Mora E, Padva L, Wuisan ZG, Liu Y, Kurz M, Marner M, Nguyen H, Amara P, Yokoyama K, Nicolet Y, Mettal U, & Schäberle TF, (2023). Genome- and metabolome-guided discovery of marine BamA inhibitors revealed a dedicated darobactin halogenase. Cell Chemical Biology, 30(8), 943–52.e7. 10.1016/j.chembiol.2023.06.011. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

