Comprehensive transcriptional analysis of ethylene and softening regulation in plums with distinct climacteric ripening behaviors
- PMID: 40652170
- PMCID: PMC12255070
- DOI: 10.1186/s12870-025-06932-w
Comprehensive transcriptional analysis of ethylene and softening regulation in plums with distinct climacteric ripening behaviors
Abstract
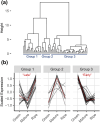
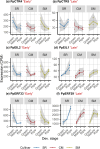
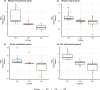
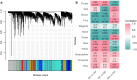
Understanding the molecular mechanisms underlying variations across climacteric categories in tree crops remains challenging, particularly due to the limitations of applying conventional genetic approaches. In this study, we examined genetically related Japanese plum (Prunus salicina Lindl.) cultivars with distinct ethylene production and responses during ripening: 'Santa Rosa' (climacteric fruit), 'Casselman' (suppressed-climacteric fruit), and 'Sweet Miriam' (non-climacteric fruit). Our primary objective was to comprehensively investigate transcriptional differences related to ethylene and fruit softening across three on-tree developmental stages: "Green" (early development), "Mature" (pre-climacteric or early climacteric), and "Ripe" (climacteric). By integrating information from Phytozome and Dicot PLAZA databases, we identified complete gene families for six ethylene-related and seven softening-related genes. Multi-developmental stage RNA-seq clustering revealed that "Late" genes, which increase in expression in ripe fruit, are associated with physiological differences among climacteric categories. We demonstrated that 'Casselman' aligns more closely with 'Sweet Miriam' than 'Santa Rosa' at the transcriptional level for these "Late" genes, consistent with their shared low ethylene production. Gene expression analyses revealed additional factors beyond ethylene, including jasmonate-related genes and NAC transcription factors as influencing climacteric ripening behavior. To extend our findings beyond the three representative cultivars, we performed qPCR on additional cultivars harvested under different field conditions and years, including 'Friar' and 'Fortune' as climacteric plums and 'Late Santa Rosa' and 'Angeleno' as suppressed-climacteric plums. PpACO1, PpPL1, PpBGAL16, PpNAC2, and PpJID1 expression patterns were conserved across cultivars and experimental conditions. Our findings provide novel insights into the transcriptional regulation of climacteric ripening and offer a strategic framework for future genetic studies in plum and other tree crops.
Keywords: Prunus salicina; Climacteric; Ethylene; Fruit softening; Non-climacteric; Suppressed climacteric; Transcriptional regulation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Hormone balance in a climacteric plum fruit and its non-climacteric bud mutant during ripening.Plant Sci. 2019 Mar;280:51-65. doi: 10.1016/j.plantsci.2018.11.001. Epub 2018 Nov 14. Plant Sci. 2019. PMID: 30824029
-
Discovery of non-climacteric and suppressed climacteric bud sport mutations originating from a climacteric Japanese plum cultivar (Prunus salicina Lindl.).Front Plant Sci. 2015 May 12;6:316. doi: 10.3389/fpls.2015.00316. eCollection 2015. Front Plant Sci. 2015. PMID: 26029222 Free PMC article.
-
Non-climacteric ripening and sorbitol homeostasis in plum fruits.Plant Sci. 2015 Feb;231:30-9. doi: 10.1016/j.plantsci.2014.11.002. Epub 2014 Nov 20. Plant Sci. 2015. PMID: 25575989
-
Structured treatment interruptions (STI) in chronic unsuppressed HIV infection in adults.Cochrane Database Syst Rev. 2006 Jul 19;2006(3):CD006148. doi: 10.1002/14651858.CD006148. Cochrane Database Syst Rev. 2006. PMID: 16856117 Free PMC article.
-
Gender differences in the context of interventions for improving health literacy in migrants: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2024 Dec 12;12(12):CD013302. doi: 10.1002/14651858.CD013302.pub2. Cochrane Database Syst Rev. 2024. PMID: 39665382
References
-
- Pech J-C, Purgatto E, Bouzayen M, Latché A. Ethylene and fruit ripening. In: Roberts J, editor. Annual Plant Reviews Online. John Wiley & Sons, Ltd; 2018. p. 275–304.
-
- Abdi N, Holford P, McGlasson WB, Mizrahi Y. Ripening behaviour and responses to propylene in four cultivars of Japanese type plums. Postharvest Biol Technol. 1997;12:21–34.
-
- Abdi N, McGlasson WB, Holford P, Williams M, Mizrahi Y. Responses of climacteric and suppressed-climacteric plums to treatment with propylene and 1-methylcyclopropene. Postharvest Biol Technol. 1998;14:29–39.

