Identity by descent and local ancestry mapping of HCV spontaneous clearance in populations of diverse ancestries
- PMID: 40652174
- PMCID: PMC12255019
- DOI: 10.1186/s12864-024-11076-6
Identity by descent and local ancestry mapping of HCV spontaneous clearance in populations of diverse ancestries
Abstract
Background: Acute infection with hepatitis C virus (HCV) affects millions of individuals worldwide. Host genetics plays a role in spontaneous clearance of the acute infection which occurs in approximately 30% of the individuals. Common variants in GPR158, genes in the interferon lambda (IFNL) cluster, and the Major Histocompatibility complex (MHC) region have been associated with HCV clearance in populations of diverse ancestry. Fine mapping of those regions has identified some key variants and amino acids as potential causal variants but the role of rare variants in those regions and in the genome, in general, has not been explored. We aimed to detect haplotypes containing rare variants related to HCV clearance using identity-by-descent (IBD) haplotype sharing between unrelated cases-case pairs and case-controls pairs in 1,739 individuals of European ancestry and 1,869 African Americans. Additionally, we aimed to detect ancestry-specific effects in African Americans using local ancestry mapping.
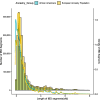
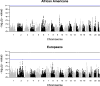
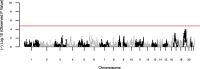
Results: We detected 2,370,341 and 1,567,748 individual pairs of IBD segments in the individuals of European ancestry and African Americans, respectively. Individuals of European descent had more segments of longer length compared to African Americans. We did not detect any significant IBD signals in the known associated or new gene regions. We also failed to detect any significant genome-wide local ancestry signals in the African Americans.
Conclusions: IBD is based on sharing of haplotypes and is most powerful in populations with a shared founder or recent common ancestor. For the complex trait of HCV clearance, we used two outbred, global populations that limited our power to detect IBD associations. Overall, in these population-based samples we failed to detect rare variations associated with HCV clearance in individuals of European ancestry and African Americans, and we didn't detect local ancestry-specific effects associated with HCV clearance in African Americans with our current sample size.
Keywords: GWAS; HCV clearance; IBD mapping; Rare variants; Unrelated individuals.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All subjects gave written informed consent for this study. Each individual study obtained consent for genetic testing from their governing Institutional Review Board (IRB) and the Johns Hopkins School of Medicine Institutional Review Board approved the overall analysis. All methods were performed with the relevant guidelines and regulations in accordance with the Declaration of Helsinki. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
Update of
-
Identity by descent mapping of HCV spontaneous clearance in populations of diverse ancestry.Res Sq [Preprint]. 2023 Jan 9:rs.3.rs-2433454. doi: 10.21203/rs.3.rs-2433454/v1. Res Sq. 2023. Update in: BMC Genomics. 2025 Jul 12;26(1):661. doi: 10.1186/s12864-024-11076-6. PMID: 36712049 Free PMC article. Updated. Preprint.
References
-
- Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, Nolt K, Nelson KE, Strathdee SA, Johnson L, Laeyendecker O, Boitnott J, Wilson LE, Vlahov D. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284(4):450–6. - PubMed
-
- Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol. 2013;10(9):553–62. - PubMed
-
- Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat. 2006;13(1):34–41. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

