Expression pattern changes of three homeologs in chemokine activity enhance antiviral response to herpesvirus infection in a newly synthesized alloheptaploid
- PMID: 40653463
- PMCID: PMC12257733
- DOI: 10.1186/s12864-025-11838-w
Expression pattern changes of three homeologs in chemokine activity enhance antiviral response to herpesvirus infection in a newly synthesized alloheptaploid
Abstract
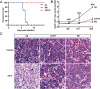
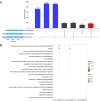
Allopolyploids often exhibit enhanced resistance to pathogen stresses. However, our understanding about the patterns that allopolyploids modify homeolog expression upon pathogen invasion remains limited. Since 2012, a disease caused by herpesvirus (CaHV) has posed a severe threat to Carassius auratus aquaculture. Therefore, the synthesis of novel allopolyploids with enhanced resistance has become one of significant priorities for its aquaculture. In this study, we first synthesized and then established a gynogenetic Carassius alloheptaploid clone (CaA7n). It possesses approximately 158 chromosomes of C. gibelio and 24 haploid chromosomes of M. amblycephala. CaHV challenge experiments showed that CaA7n inherited high resistance from its paternal M. amblycephalus, exhibiting a 100% survival rate after CaHV infection. Subsequently, we revealed distinct transcriptional responses among CaA7n and its parents to CaHV infection and identified two key modules. The egiengenes in the module that positively correlated with CaA7n resistance were mainly enriched in chemokine activity GO terms. Finally, we described a profound expression alteration of three homeologs in CaA7n, including additive and non-additive expression patterns. After CaHV infection, three homeologs mainly involved in chemokine activity changed their expression patterns in CaA7n. Moreover, homeologs derived from M. amblycephala associated with chemokine activity, which showed altered expression levels, may enhance the antiviral immune response of CaA7n. This study not only establishes CaA7n as a promising CaHV-resistant candidate for aquaculture but also elucidates how allopolyploids reconfigure parental homeolog expression networks to enhance antiviral defenses, advancing our understanding of allopolyploid adaptation mechanisms under pathogenic pressure.
Keywords: Carassius; Allopolyploid; Antiviral response; Chemokine; Herpesvirus; Homeologs.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Gui JF, Zhou L, Li XY. Rethinking fish biology and biotechnologies in the challenge era for burgeoning genome resources and strengthening food security. Water Biol Secur. 2022;1(1):100002. 10.1016/j.watbs.2021.11.001. - DOI
-
- Gui JF. Chinese wisdom and modern innovation of aquaculture. Water Biol Secur. 2024;3(3):100271. 10.1016/j.watbs.2024.100271. - DOI
-
- Zhou L, Gui JF. Natural and artificial polyploids in aquaculture. Aquacult Fish. 2017;2(3):103–11. 10.1016/j.aaf.2017.04.003. - DOI
-
- Zhou L, Gui JF. Epigenetics in hybridization and polyploidization of aquatic animals. In: Piferrer F, Wang HP, eds. Epigenetics in Aquaculture. Wiley. 2023;287–99.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

