Multi-omics analysis for identifying cell-type-specific and bulk-level druggable targets in Alzheimer's disease
- PMID: 40653482
- PMCID: PMC12257706
- DOI: 10.1186/s12967-025-06739-1
Multi-omics analysis for identifying cell-type-specific and bulk-level druggable targets in Alzheimer's disease
Abstract
Background: Analyzing disease-linked genetic variants via expression quantitative trait loci (eQTLs) helps identify potential disease-causing genes. Previous research prioritized genes by integrating Genome-Wide Association Study (GWAS) results with tissue-level eQTLs. Recent studies have explored brain cell type-specific eQTLs, but a systematic analysis across multiple Alzheimer's disease (AD) genome-wide association study (GWAS) datasets or comparisons between tissue-level and cell type-specific effects remain limited. Here, we integrated brain cell type-level and bulk-level eQTL datasets with AD GWAS datasets to identify potential causal genes.
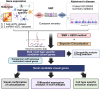
Methods: We used Summary Data-Based Mendelian Randomization (SMR) and Bayesian Colocalization (COLOC) to integrate AD GWAS summary statistics with eQTLs datasets. Combining data from five AD GWAS, two single-cell eQTL datasets, and one bulk eQTL dataset, we identified novel candidate causal genes and further confirmed known ones. We investigated gene regulation through enhancer activity using H3K27ac and ATAC-seq data, performed protein-protein interaction (PPI) and pathway enrichment, and conducted a drug/compound enrichment analysis with Drug Signatures Database (DSigDB) to support drug repurposing for AD.
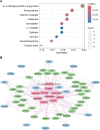
Results: We identified 28 candidate causal genes for AD, of which 12 were uniquely detected at the cell-type level, 9 were exclusive to the bulk level and 7 detected in both. Among the 19 cell-type level candidate causal genes, microglia contributed the highest number of candidate genes, followed by excitatory neurons, astrocytes, inhibitory neurons, oligodendrocytes, and oligodendrocyte precursor cells (OPCs). PABPC1 emerged as a novel candidate causal gene in astrocytes. We generated PPI networks for the candidate causal genes and found that pathways such as membrane organization, cell migration, and ERK1/2 and PI3K/AKT signaling were enriched. The AD-risk variant associated with candidate causal gene PABPC1 is located near or within enhancers only active in astrocytes. We classified the 28 genes into three drug tiers and identified druggable interactions, with imatinib mesylate emerging as a key candidate. A drug-target gene network was created to explore potential drug targets for AD.
Conclusions: We systematically prioritized AD candidate causal genes based on cell type-level and bulk level molecular evidence. The integrative approach enhances our understanding of molecular mechanisms of AD-related genetic variants and facilitates interpretation of AD GWAS results.
Keywords: Alzheimer’s disease; Astrocytes; Causal genes; Cell type; Drug repurposing; GWAS; Gene expression; Genetic variant; SNP; eQTL.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study used publicly available datasets, no ethics approval and consent to participate was not required. Consent for publication: This study used publicly available datasets. All data were anonymized, and no information that could reveal the identity of participants was used. Therefore, consent for publication from individual participants was not required. Competing interests: A.S. has received support from Avid Radiopharmaceuticals, a subsidiary of Eli Lilly (in kind contribution of PET tracer precursor) and participated in Scientific Advisory Boards (Bayer Oncology, Eisai, Novo Nordisk, and Siemens Medical Solutions USA, Inc) and an Observational Study Monitoring Board (MESA, NIH NHLBI), as well as several other NIA External Advisory Committees. He also serves as Editor-in-Chief of Brain Imaging and Behavior, a Springer-Nature Journal. S. L., T. R., P. B., D. C., D. B., N. T., K. N., S. C., M. C., Y. H., and T. P. have no interest to declare. The funders had no role in the study's design, the collection, analyses, or interpretation of data, the writing of the manuscript, or the decision to publish the results.
Figures




Update of
-
Multi-Omics Analysis for Identifying Cell-Type-Specific Druggable Targets in Alzheimer's Disease.medRxiv [Preprint]. 2025 Jan 9:2025.01.08.25320199. doi: 10.1101/2025.01.08.25320199. medRxiv. 2025. Update in: J Transl Med. 2025 Jul 13;23(1):788. doi: 10.1186/s12967-025-06739-1. PMID: 39830273 Free PMC article. Updated. Preprint.
References
-
- Tanzi RE, Bertram L. New frontiers in Alzheimer’s disease genetics. Neuron. 2001;32(2):181–4. - PubMed
-
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–3. - PubMed
MeSH terms
Grants and funding
- R01 AG019771/AG/NIA NIH HHS/United States
- U19 AG074879/NH/NIH HHS/United States
- U19 AG0748790/NH/NIH HHS/United States
- CLEAR-AD Scholarship from U19 AG074879/NH/NIH HHS/United States
- Alzheimer's Disease Neuroimaging Initiative Health Enhancement Scientific Program (ADNI HESP) subaward from U19 AG024904/NH/NIH HHS/United States
- R01 LM012535/NH/NIH HHS/United States
- R01 AG057739/NH/NIH HHS/United States
- R01 LM013463/LM/NLM NIH HHS/United States
- U01 AG072177/NH/NIH HHS/United States
- U01 AG068057/NH/NIH HHS/United States
- T32 AG071444/AG/NIA NIH HHS/United States
- U01 AG072177/AG/NIA NIH HHS/United States
- P30 AG010133/AG/NIA NIH HHS/United States
- U19 AG074879/AG/NIA NIH HHS/United States
- R01 AG068193/NH/NIH HHS/United States
- T32 AG071444/NH/NIH HHS/United States
- U01 AG068057/AG/NIA NIH HHS/United States
- U19 AG024904/NH/NIH HHS/United States
- P30 AG010133/NH/NIH HHS/United States
- U19 AG024904/AG/NIA NIH HHS/United States
- R01 AG019771/NH/NIH HHS/United States
- P30 AG072976/NH/NIH HHS/United States
- R01 AG057739/AG/NIA NIH HHS/United States
- R01 AG068193/AG/NIA NIH HHS/United States
- R01 LM012535/LM/NLM NIH HHS/United States
- R01 LM013463/NH/NIH HHS/United States
- P30 AG072976/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

