Lymphoid and CXCR4 Cell Targeted Lipid Nanoparticles Facilitate HIV-1 Proviral DNA Excision
- PMID: 40653915
- PMCID: PMC12477575
- DOI: 10.1002/adhm.202501190
Lymphoid and CXCR4 Cell Targeted Lipid Nanoparticles Facilitate HIV-1 Proviral DNA Excision
Abstract
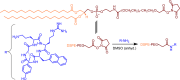
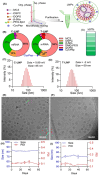
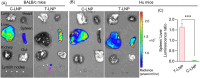
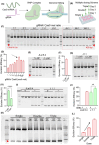
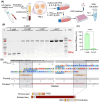
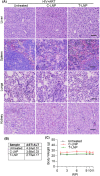
Advancements in antiretroviral therapy (ART) enable those living with the human immunodeficiency virus type one (HIV-1) to lead longer, healthier lives free from disease comorbidities. However, lifelong ART poses challenges. These include social stigma, medication costs, drug accessibility, mental health, and drug-related toxicities. Moreover, ART does not eliminate latent HIV-1 DNA. Viral persistence in tissue and cell reservoirs results in viral rebound after ART interruption. New strategies are required to achieve a functional HIV-1 cure. To excise latent HIV-1, C-X-C motif chemokine receptor 4 (CXCR4) ligand-decorated lymphoid tissue-targeting lipid nanoparticles (LNPs) for CRISPR-Cas9/gRNA delivery are developed. These LNPs enhance mRNA translation and demonstrate CXCR4-mediated improved uptake to eliminate HIV-1 DNA in infected CD4+ T cells. LNPs also facilitate targeted drug delivery, achieving HIV-1 DNA excision in ART-treated, infected humanized mice. This study emphasizes the potential of tissue and cell-targeted LNPs for effective HIV-1 DNA excision.
Keywords: CRISPR; HIV‐1 infection; excision of HIV‐1 DNA; lipid nanoparticles; targeted mRNA delivery.
© 2025 The Author(s). Advanced Healthcare Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare that Dr. Howard Gendelman is co‐founder of Exavir Therapeutics, Inc. The biotechnology company is developing ultra‐long‐acting drugs. The drugs in development are not linked to those created in the current report. All other authors declare no competing interests.
Figures


References
-
- WHO, HIV Factsheets , https://www.who.int/news‐room/fact‐sheets/detail/hiv‐aids, accessed.
-
- Koethe J. R., Lagathu C., Lake J. E., Domingo P., Calmy A., Falutz J., Brown T. T., Capeau J., Nat. Rev. Dis. Primers 2020, 6, 48. - PubMed
-
- Chun T.‐W., Finzi D., Margolick J., Chadwick K., Schwartz D., Siliciano R. F., Nat. Med. 1995, 1, 1284. - PubMed
-
- Chun T.‐W., Carruth L., Finzi D., Shen X., DiGiuseppe J. A., Taylor H., Hermankova M., Chadwick K., Margolick J., Quinn T. C., Kuo Y.‐H., Brookmeyer R., Zeiger M. A., Barditch‐Crovo P., Siliciano R. F., Nature 1997, 387, 183. - PubMed
-
- Neumann A. U., Tubiana R., Calvez V., Robert C., Li T.‐S., Agut H., Autran B., Katlama C., AIDS 1999, 13, 677. - PubMed
MeSH terms
Substances
Grants and funding
- R01 NS036126/NS/NINDS NIH HHS/United States
- R01 NS126089/NH/NIH HHS/United States
- R01 DA054535/NH/NIH HHS/United States
- P30 MH062261/MH/NIMH NIH HHS/United States
- R01 NS36126/NH/NIH HHS/United States
- 2R01 NS034239/NH/NIH HHS/United States
- 5R01MH115860/MH/NIMH NIH HHS/United States
- P01 MH064570/MH/NIMH NIH HHS/United States
- DP1 DA053719/DA/NIDA NIH HHS/United States
- PO1 MH64570/NH/NIH HHS/United States
- R01 NS126089/NS/NINDS NIH HHS/United States
- P30 MH062261/NH/NIH HHS/United States
- R01 MH115860/MH/NIMH NIH HHS/United States
- 5R01MH121402/NH/NIH HHS/United States
- National Institute of Allergy and Infectious Diseases
- PO1 DA028555/NH/NIH HHS/United States
- P01 DA028555/DA/NIDA NIH HHS/United States
- R01 DA054535/DA/NIDA NIH HHS/United States
- 1R01Al158160/NH/NIH HHS/United States
- R01 MH121402/MH/NIMH NIH HHS/United States
- R01NS126089/NS/NINDS NIH HHS/United States
- R01 NS034239/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

