This is a preprint.
Transcription Factor (TF) validation using Dam-IT simultaneously captures genome-wide TF-DNA binding, direct gene regulation, and chromatin accessibility in plant cells
- PMID: 40654631
- PMCID: PMC12248053
- DOI: 10.1101/2025.05.06.652526
Transcription Factor (TF) validation using Dam-IT simultaneously captures genome-wide TF-DNA binding, direct gene regulation, and chromatin accessibility in plant cells
Abstract
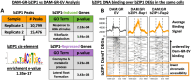
Transcription Factors (TFs) govern vast networks of gene regulation. However, TF-DNA binding and TF-gene regulation datasets are typically measured separately due to experimental constraints, making it challenging to disentangle true biological relationships from batch effects. To fill this gap, we developed DamID-seq Incorporating Transcriptomics (Dam-IT), which simultaneously captures TF-DNA binding, direct TF-gene regulation, and chromatin accessibility in the same batch of cells. Dam-IT uses a transient cell-based TF-target validation system that is scalable and flexible to many experimental designs. As proof of concept, we used Dam-IT to reveal that bZIP1 directly regulates genes by binding to DNA regions of relatively low chromatin accessibility, supporting a "Hit-and-Run" mechanism of transcription.
Keywords: DNA binding; DamID; chromatin accessibility; gene regulation; transcription factor.
Figures

References
-
- O'Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 2016. May 19;165(5):1280–1292. doi: 10.1016/j.cell.2016.04.038. Erratum in: Cell. 2016 Sep 8;166(6):1598. doi: 10.1016/j.cell.2016.08.063. - DOI - PMC - PubMed
-
- Tao XY, Guan XY, Hong GJ, He YQ, Li SJ, Feng SL, Wang J, Chen G, Xu F, Wang JW, Xu SC. Biotinylated Tn5 transposase-mediated CUT&Tag efficiently profiles transcription factor-DNA interactions in plants. Plant Biotechnol J. 2023. Jun;21(6):1191–1205. doi: 10.1111/pbi.14029. Epub 2023 Mar 2. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
