This is a preprint.
Comparative functional and evolutionary analysis of essential germline stem cell genes across the genus Drosophila and two outgroup species
- PMID: 40654705
- PMCID: PMC12247945
- DOI: 10.1101/2025.04.30.651540
Comparative functional and evolutionary analysis of essential germline stem cell genes across the genus Drosophila and two outgroup species
Abstract
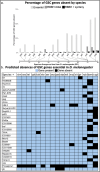
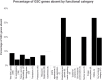
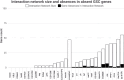
In Drosophila melanogaster, bag of marbles (bam) encodes a protein essential for germline stem cell daughter (GSC) differentiation in early gametogenesis. Despite its essential role in D. melanogaster, direct functional evaluation of bam in other closely related Drosophila species reveal this essential function is not necessarily conserved. In D. teissieri, for example, bam is not essential for GSC daughter differentiation. Here, we generated bam null alleles using CRISPR-Cas9 in a species more distantly related to D. melanogaster, D. americana, to interrogate whether bam's essential GSC differentiation function is novel to the melanogaster species group or a function more basal to the Drosophila genus. To further characterize the extent of the functional flexibility of other GSC regulating genes, we generated a gene ortholog dataset for 366 GSC regulating genes essential in D. melanogaster across 15 additional Drosophila and two outgroup species. We find that bam's essential GSC function is conserved between D. melanogaster and D. americana and therefore originated prior to the formation of the melanogaster species group. Additionally, we find that ~8% of the 366 GSC genes essential in D. melanogaster are absent in at least one of the 17 species in our ortholog dataset. These results indicate that developmental systems drift (DSD), in which the specific genes regulating a function may change, but the final phenotype is retained, occurs in stem cell regulation and the production of gametes across Drosophila species.
Keywords: CRISPR; bam; comparative functional analysis; germline stem cells.
Figures


Similar articles
-
Functional Divergence of the bag-of-marbles Gene in the Drosophila melanogaster Species Group.Mol Biol Evol. 2022 Jul 2;39(7):msac137. doi: 10.1093/molbev/msac137. Mol Biol Evol. 2022. PMID: 35714266 Free PMC article.
-
Female-germline specific protein Sakura interacts with Otu and is crucial for germline stem cell renewal and differentiation and oogenesis.Elife. 2025 Jul 15;13:RP103828. doi: 10.7554/eLife.103828. Elife. 2025. PMID: 40663062 Free PMC article.
-
Stress-induced Cdk5 activity enhances cytoprotective basal autophagy in Drosophila melanogaster by phosphorylating acinus at serine437.Elife. 2017 Dec 11;6:e30760. doi: 10.7554/eLife.30760. Elife. 2017. PMID: 29227247 Free PMC article.
-
Bioengineered nerve conduits and wraps for peripheral nerve repair of the upper limb.Cochrane Database Syst Rev. 2022 Dec 7;12(12):CD012574. doi: 10.1002/14651858.CD012574.pub2. Cochrane Database Syst Rev. 2022. PMID: 36477774 Free PMC article.
-
Nivolumab for adults with Hodgkin's lymphoma (a rapid review using the software RobotReviewer).Cochrane Database Syst Rev. 2018 Jul 12;7(7):CD012556. doi: 10.1002/14651858.CD012556.pub2. Cochrane Database Syst Rev. 2018. PMID: 30001476 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
