This is a preprint.
High-Sensitive Spatial Proteomics for Pancreatic Cancer Progression Analysis
- PMID: 40654937
- PMCID: PMC12247709
- DOI: 10.1101/2025.05.01.651678
High-Sensitive Spatial Proteomics for Pancreatic Cancer Progression Analysis
Abstract
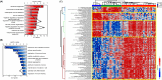
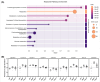
Pancreatic cancer remains as one of the most challenging malignancies to diagnose and treat due to the late development of symptoms and limited early diagnostic options. Intraductal papillary mucinous neoplasms (IPMNs) are non-invasive precursors to invasive pancreatic ductal adenocarcinoma (PDAC)and an understanding of the changes in patterns of protein expression that accompany the progression from normal ductal (ND) cell, to IPMN to PDAC may provide avenues for improved earlier detection. In this study, we present an optimized spatial tissue proteomics workflow, termed SP-Max (Spatial Proteomics Optimized for Maximum Sensitivity and Reproducibility in Minimal Sample), designed to maximize protein recovery and quantification from limited laser micro dissected (LMD) samples. Our workflow enabled the identification of more than 6,000 proteins and the quantification of over 5,200 protein groups from FFPE tissue contours of pancreatic tissues. Comparative analyses across ND, IPMN, and PDAC revealed critical molecular differences in protein pathways and potential markers of progression. SP-Max provides a systematic, reproducible approach that significantly enhances our ability to study precancerous lesions and cancer progression in pancreatic tissues at unprecedented resolution.
Keywords: FFPE Tissue; High-sensitivity Mass Spectrometry; Intraductal Papillary Mucinous Neoplasm; Pancreatic Ductal Adenocarcinoma; Spatial Proteomics.
Conflict of interest statement
Conflict of Interest H.Z. discloses serving as a co-founder of Complete Omics Inc. The remaining authors have no conflicts of interest to declare.
Figures



References
-
- Rahib L.; Smith B. D.; Aizenberg R.; Rosenzweig A. B.; Fleshman J. M.; Matrisian L. M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014, 74 (11), 2913–2921. DOI: 10.1158/0008-5472.CAN-14-0155 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
