This is a preprint.
Identification and Overexpression of Endogenous Transcription Factors to Enhance Lipid Accumulation in the Commercially Relevant Species Chlamydomonas pacifica
- PMID: 40655021
- PMCID: PMC12247862
- DOI: 10.1101/2025.05.01.651737
Identification and Overexpression of Endogenous Transcription Factors to Enhance Lipid Accumulation in the Commercially Relevant Species Chlamydomonas pacifica
Abstract
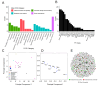
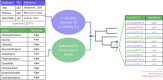
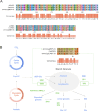
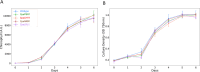
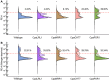
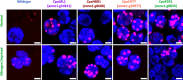
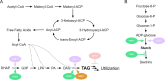
Sustainable low-carbon energy solutions are critical to mitigating global carbon emissions. Algae-based platforms offer potential by converting carbon dioxide into valuable products while aiding carbon sequestration. However, scaling algae cultivation faces challenges like contamination in outdoor systems. Previously, our lab evolved Chlamydomonas pacifica, an extremophile green alga, which tolerates high temperature, pH, salinity, and light, making it ideal for large-scale bioproduct production, including biodiesel. Here, we enhanced lipid accumulation in evolved C. pacifica by identifying and overexpressing key endogenous transcription factors through genome-wide in-silico analysis and in-vivo testing. These factors include Lipid Remodeling Regulator 1 (CpaLRL1), Nitrogen Response Regulator 1 (CpaNRR1), Compromised Hydrolysis of Triacylglycerols 7 (CpaCHT7), and Phosphorus Starvation Response 1 (CpaPSR1). Under nitrogen deprivation, CpaLRL1, CpaNRR1, and CpaCHT7 overexpression enhanced lipid accumulation compared to wildtype. However, CpaPSR1 increased lipid accumulation compared to wildtype in normal media despite causing no effect under nitrogen depravation, highlighting the difference in function based on media conditions. Notably, lipid analysis of CpaPSR1 under normal media conditions revealed a 2.4-fold increase in triglycerides (TAGs) compared to the wild type, highlighting its potential for biodiesel production. This approach provides a framework for transcription factor-focused metabolic engineering in algae, advancing bioenergy and biomaterial production.
Keywords: biofuels; biotechnology; computational modeling; microalgae; nitrogen-deprivation; sustainability; transcription factors.
Conflict of interest statement
Ethics declarations Competing Interests SM and MDB are co-founders of and hold equity in Algenesis Inc., a company that could potentially benefit from this research. MT is an employee and shareholder in Algenesis Inc. The remaining authors declare that their research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Stern D. I. The role of energy in economic growth. Ann. N. Y. Acad. Sci. 1219, 26–51 (2011). - PubMed
-
- Johnsson F., Kjärstad J. & Rootzén J. The threat to climate change mitigation posed by the abundance of fossil fuels. Clim. Policy 19, 258–274 (2019).
-
- Olabi A. G. & Abdelkareem M. A. Renewable energy and climate change. Renew. Sustain. Energy Rev. 158, 112111 (2022).
-
- Capellán-Pérez I., Mediavilla M., de Castro C., Carpintero Ó. & Miguel L. J. Fossil fuel depletion and socio-economic scenarios: An integrated approach. Energy 77, 641–666 (2014).
-
- Barbosa M. J., Janssen M., Südfeld C., D’Adamo S. & Wijffels R. H. Hypes, hopes, and the way forward for microalgal biotechnology. Trends Biotechnol. 41, 452–471 (2023). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
